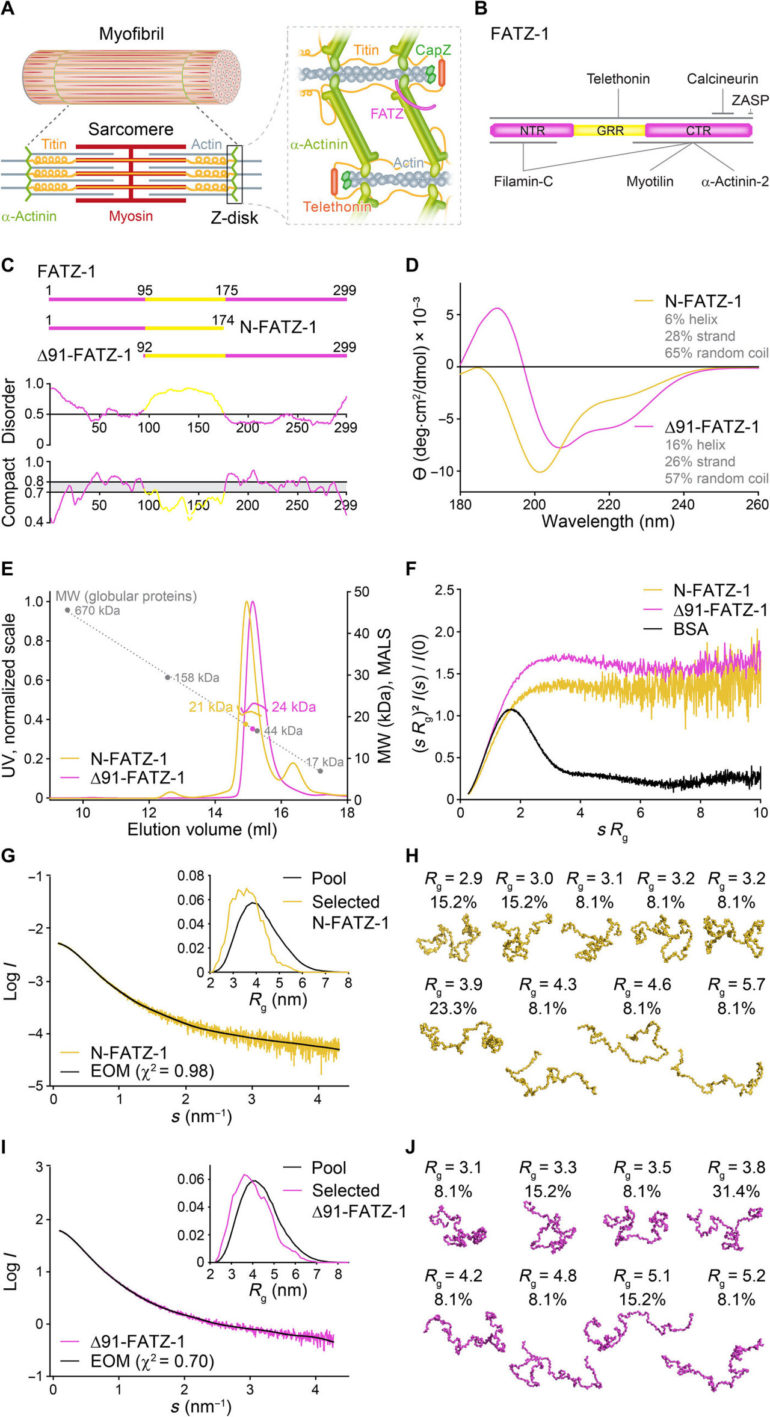
Alpha-actinin can cross-link actin filaments and anchor them to the Z-disk in sarcomeres. Sarcomeres are a structural unit of myofibril in striated muscle. The FATZ (filamin, α-actinin- and telethonin-binding protein of the Z-disk) protein can interact with α-actinin and other core Z-disk proteins that contribute to myofibril assembly and maintenance. In a new report now on Science Advances, Antonio Sponga and an international research team in Austria, Germany, Russia, Poland and the U.K. detailed the first structure and cellular validation of the α-actinin-2 complex with a Z-disk partner, FATZ-1, to form a conformal ensemble. The FATZ-1 formed a tight fuzzy complex with α-actinin-2 with a proposed interaction mechanism via molecular recognition elements and secondary binding sites. The obtained integrative model revealed a polar architecture of the complex in combination with the FATZ-1 multivalent scaffold function to organize interaction partners and stabilize.
Sarcomere
The contracting muscles can regulate voluntary animal movement and involuntary heart beating, and sarcomeres are the basic contractile units of striated muscle cells. They are composed of arrays of thin (actin) and thick (myosin) filaments that slide past each other during contraction. The Z-disk can form the boundary between adjacent sarcomeres, where anti-parallel actin filaments are anchored. A suitably stable anchoring structure must be generated by the interaction between myosin and actin. The Z-disk can fulfill this role by acting as a mechanical hub and a signaling platform to allow the transmission of tension during contraction and the duration and transmission of information of biomechanical stress. As a result, any mutations that disrupt the Z-disk architecture and function could risk causing skeletal and cardiac dysfunction.
The protein complex
Alpha-actinin is an F-actin cross-linking protein in muscle Z-disks, which forms a major Z-disk component that cross-links antiparallel actin filaments from adjacent sarcomeres to serve as a binding platform for multiple Z-disk proteins, including FATZ-1. The FATZ proteins can bind to α-actinin through their c-terminal region and to domains of the Enigma family members via a specific c-terminal recognition motif. In this work, Antonio Sponga et al. demonstrated how FATZ proteins contained intrinsically disordered regions (IDRs) best described as a conformational ensemble, which are less stable and lack a stable tertiary structure. In addition to biophysical characterization methods, the team used X-ray crystallography and small-angle X-ray scattering to describe a “fuzzy” α-actinin-2/FATZ-1 complex. The FATZ-1 protein can play an organizational role in the Z-disk due to its multivalent scaffolding properties and form a tight complex of polar architecture with α-actinin-2.
The FATZ protein family
The FATZ protein family are found across all vertebrates where human FATZ-1, FATZ-2, and FATZ-3 share 34 to 40 percent sequence identity. The scientists recognized proteolysis-resistant fragments, after conducting proteolysis experiments. When they combined size exclusion chromatography (SEC) with multiangle light scattering, they noted the predominant monomers under experimental conditions. They then further characterized the monomers using SEC combined with small angle X-ray scattering and also highlighted the intrinsically disordered/ensemble-state nature of the monomers using single-quantum coherence (HSQC) spectra, for both constructs. To understand the binding stoichiometry of the FATZ-1-to-3 proteins to α-actinin-2, Sponga et al. used size exclusion chromatography-multiangle light scattering (SEC-MALS). To characterize the binding stoichiometry of the FATZ-1-to-3 proteins to α-actinin-2, Sponga et al. used SEC-MALS. The outcome showed how each of the three FATZ proteins formed a tight complex with α-actinin-2, with a binding stoichiometry of two FATZ molecules per α-actinin-2 dimer. That is one FATZ molecule per α-actinin-2 subunit. The team next used isothermal titration calorimetry (ITC) to quantify the interaction affinity.
![Crystal structures of α-actinin-2/FATZ-1 reveal two linear binding motifs in FATZ-1. (A) Crystal structure of rod-α-actinin-2/mini-FATZ-1 (in green/magenta), along with the determined FATZ-1 consensus sequence (35 to 80% and 60 to 84% pairwise sequence identity for LM1 and LM2, respectively). Cross-linked residues are indicated by blue, red, and gray stars/balls/sticks on the sequence/structure. Identified Se-Mets are shown in yellow. The rod-α-actinin-2 dimer is assembled through a crystallographic twofold axis between symmetry mates (black circle). Interacting residues (rod-α-actinin-2 in italics), along with helices from SR1/SR2 (h1, h2, and h3) and SR3/SR4 (h1′, h2′, and h3′), are shown in close-up views. (B) Crystal structure of hd-α-actinin-2/Δ91-FATZ-1 (LM1 and LM2 as magenta cartoon and transparent gray surface; hd-α-actinin-2 color-coded as in Fig. 2A). (C) Comparison of unbound [Protein Data Bank (PDB) code 4D1E] and bound (this work) hd-α-actinin-2. ABD and EF1-2 of unbound hd-α-actinin-2 are shown with transparency. ITC analysis for the interaction of LM1 peptide with α-actinin-2 (D), LM2 peptide with α-actinin-2 (E), and RRE Δ91-FATZ-1 mutant with α-actinin-2 (F). SEC-MALS analysis for the interaction of RRE Δ91-FATZ-1 mutant with rod-α-actinin-2 (G) and Δ91-FATZ-1 with E. histolytica rod-α-actinin-2 (H). Science Advances, doi: 10.1126/sciadv.abg7653 Order from disorder in the sarcomere](https://techandsciencepost.com/wp-content/uploads/2021/06/1624197113_437_Order-from-disorder-in-the-sarcomere.jpg)
Multiple binding sites for the protein complex
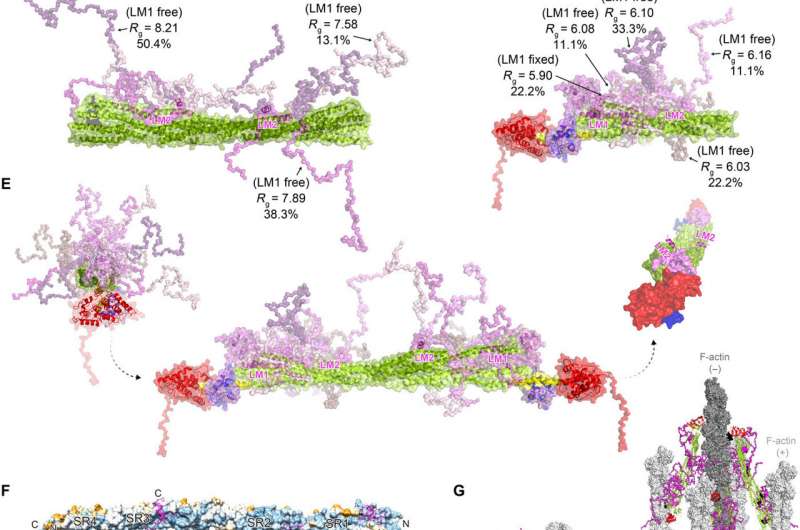
The team noted how FATZ-1 interacted with α-actinin-2 via multiple binding sites. To narrow down the FATZ-1 binding sites, Sponga et al. used limited proteolysis and chemical cross-linking coupled with mass spectrometry on the protein complex. To then aid the crystallization of this protein complex, the team then also combined the information from the peptide array and generated a shorter construct known as mini-FATZ-1 for further studies on their structural biology. The scientists then validated the fuzzy models developed in the work using calculated and experimentally derived intrinsic viscosity—a hydrodynamic parameter of protein conformation. To then understand the contribution of α-actinin-2 to localize FATZ proteins onto the Z-disk of the sarcomere, Sponga et al. transfected GFP-tagged FATZ-1 or FATZ-2 proteins into immortalized mouse myoblasts or neonatal rat cardiomyocytes. Both FATZ-1 and -2 proteins correctly targeted the Z-disk and co-localized with α-actinin-2.
Outlook
In this way, Antonio Sponga and colleagues described how the sarcomere assembly started from Z-bodies of α-actinin-2, to include proteins such as FATZ, myotilin, and actin, to name a few. The outcome indicates that proteins of the FATZ family are available in Z-bodies and mature Z-disks with a role in protein signaling pathways to bind calcineurin. The team highlighted the role of FATZ-1, the most studied family member and its interaction with the major Z-disk protein α-actinin-2. The structure and binding mechanism of the fuzzy α-actinin-2/FATZ-1 complex supported FATZ-1 function as a classical scaffolding protein in the Z-disk assembly. Further studies will reveal if the same principles apply under physiological conditions in living cells.
How do our muscles work? Scientists reveal important new insights into muscle protein
More information:
Sponga A. et al. Order from disorder in the sarcomere: FATZ forms a fuzzy but tight complex and phase-separated condensates with α-actinin, Science Advances, DOI: 10.1126/sciadv.abg7653
Chambers M. C. et al. A cross-platform toolkit for mass spectrometry and proteomics. Nature Biotechnology, doi.org/10.1038/nbt.2377
Salman F. Banani et al, Biomolecular condensates: organizers of cellular biochemistry, Nature Reviews Molecular Cell Biology (2017). DOI: 10.1038/nrm.2017.7
2021 Science X Network
Citation:
Order from disorder in the sarcomere (2021, June 18)
retrieved 20 June 2021
from https://phys.org/news/2021-06-disorder-sarcomere.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.