Many patients with pancreatic cancer have only about a 10% chance of survival within five years of their diagnosis because they tend to become resistant to chemotherapy, past studies have indicated.
A “time machine” that Purdue University engineers designed to observe pancreatic cancer behavior over time suggests a new drug testing approach that could help scientists better catch resistance.
The researchers found that testing potential drugs on multiple tumor cell subtypes—rather than on just one cell subtype—can reveal drug resistance that may occur due to how different cancer subtypes interact with each other.
The study was recently published in the Royal Society of Chemistry journal Lab on a Chip.
“The drug discovery and screening process has been using one cancer cell subtype and studying how it interacts with neighboring non-cancer cells, but this may overestimate the efficacy of the drug,” said Bumsoo Han, a Purdue professor of mechanical engineering and program leader of the Purdue Center for Cancer Research. Han has a courtesy appointment in biomedical engineering.
“By condensing time to look at how cancer cells interact within a pancreatic tumor, we found that one cancer cell subtype can not only be more drug-resistant than the others, but drug-sensitive cells can also become resistant through interaction between the subtypes.”
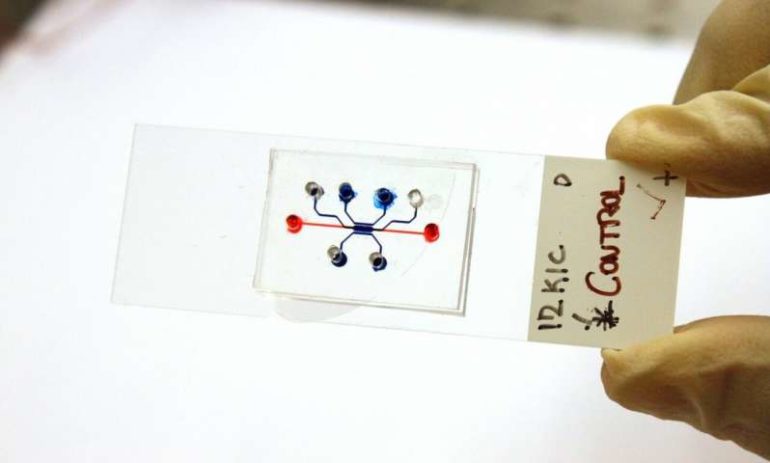
The “time machine” is a type of lab tool called a microfluidic device. These devices are gum strip-sized platforms, such as a chip or slide, where cancer cells can be cultured in channels smaller than a millimeter in diameter. The cells then grow in a lifelike environment on the platform, such as in a collagen tube that Han’s lab created to mimic the pancreatic duct.
Microfluidic devices are starting to become more mainstream in the drug development process because they allow scientists to test drugs in realistic simulations of a biological system using real tissue samples, but on a faster time scale than in animal models.
Han’s group found that about 25% of 2019 research publications indexed by PubMed, a biomedical literature database, had used microfluidic devices as models to study tumors from animals or patients.
But most microfluidic devices just show late-stage tumor growth. With Han’s device, scientists can load in cell lines from an animal model or patient before gene mutation has happened, enabling them to see all stages of tumor progression.
While findings made using microfluidic devices need to be validated in humans before being put into clinical practice, they can still shorten the drug development process by offering new research approaches.
The findings from Han’s device highlight the need for studying interactions between cancer cells.
“Not much research has been done on what kind of interaction happens within tumors, so those mechanisms of drug resistance have been overlooked,” Han said.
These findings are already informing the development of new drug compounds.
Zhong-Yin Zhang, the director of the Purdue Institute for Drug Discovery, is using Han’s microfluidic device to test a compound aimed at blocking an oncogenic process that Zhang’s lab has previously identified as playing a role in cancer development.
The device allows Zhang’s team to evaluate the compound not only for pancreatic cancer specifically, but also on multiple cancer cell subtypes.
“The nice thing about this device is that we don’t have to use as much of a compound to see how well it works,” said Zhang, who is also a distinguished professor of medicinal chemistry in Purdue’s Department of Medicinal Chemistry and Molecular Pharmacology and Department of Chemistry.
Pancreatic cancer ‘time machine’ exposes plot twist in cell growth and invasion
More information:
Hye-ran Moon et al, An engineered pancreatic cancer model with intra-tumoral heterogeneity of driver mutations, Lab on a Chip (2020). DOI: 10.1039/D0LC00707B
Citation:
‘Time machine’ offers new pancreatic cancer drug testing approach (2020, October 29)
retrieved 29 October 2020
from https://phys.org/news/2020-10-machine-pancreatic-cancer-drug-approach.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.