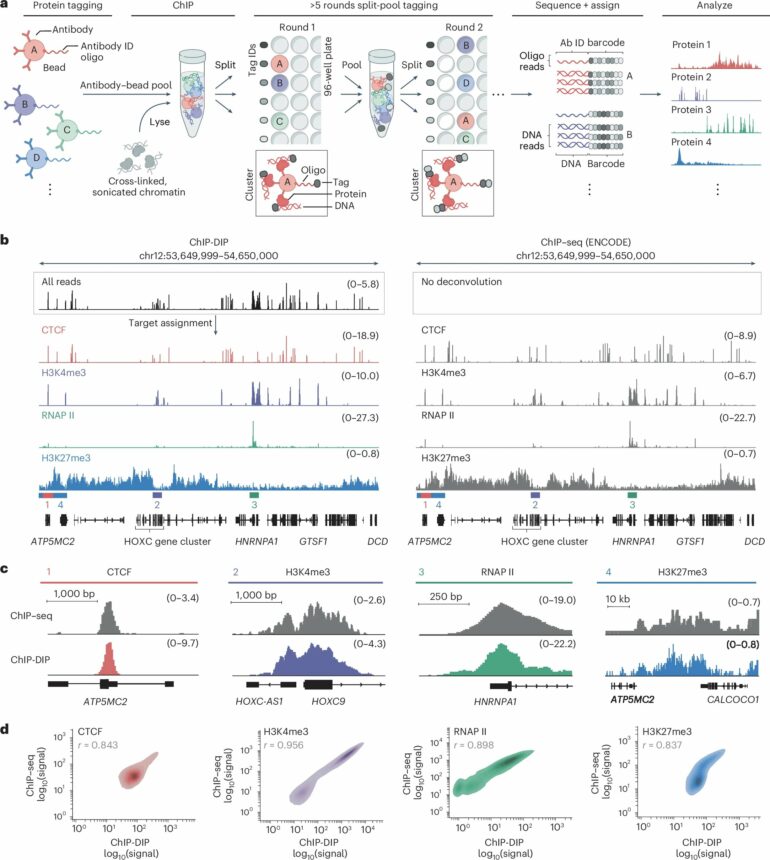
Caltech researchers have developed a new method to map the positions of hundreds of DNA-associated proteins within cell nuclei all at the same time. The method, called ChIP–DIP (Chromatin ImmunoPrecipitation Done In Parallel), is a versatile tool for understanding the inner workings of the nucleus during different contexts, such as disease or development.
The research was conducted in the laboratory of Mitchell Guttman, professor of biology, and is described in a paper that appears in the journal Nature Genetics.
Nearly all cells in the human body contain the same DNA, which encodes the blueprint for creating every cell type in the body and directing their activities. Despite having the same genetic material, different cell types express unique sets of proteins, allowing for the various cells to perform their specialized functions and to adapt to conditions within their environments. This is possible because of careful regulation within the nucleus of each cell and involves thousands of regulatory proteins that localize to precise places in the nucleus.
Because the nucleus is 50 times smaller than the width of a human hair, a cell’s DNA—which measures 2 meters long when stretched out end to end—is wrapped up like spools of thread around protein structures called histones. Histones can be modified and reorganized during a cell’s lifetime to reveal different sections of DNA while packing others away, allowing the cell to change the set of proteins that are expressed and, hence, its function. (As an analogy, imagine packing away your winter clothes in the back of your closet during summer and bringing them back out as it gets colder.)
Thus, though the full complement of DNA within a brain cell and a liver cell is the same, the two cells have different histone modifications and other regulatory proteins, allowing the expression of unique sets of genes in each. If this regulation goes awry, it can lead to serious diseases such as cancer, inflammatory diseases, or neurodegeneration.
Despite its importance, understanding gene regulation has been very challenging because previous methods to study regulatory proteins map them one at a time. ChIP–DIP now enables researchers to simultaneously map hundreds of DNA-associated regulatory proteins and take snapshots of how they change over time.
Isabel Goronzy (Ph.D. ’24), a former graduate student in the Guttman lab and co-first author of the new paper, explains the power of the new technique, “We used ChIP–DIP to show how after an inflammatory event, cells of the immune system rapidly alter their histone proteins within a span of hours in order to activate inflammatory genes.
“We also used ChIP–DIP to identify combinations of proteins that regulate which genes are active or will become active in response to stress or during development. Previous consortium-based international projects have taken nearly a decade to conduct a few thousand experiments, but we have now done over 500 in the span of a few weeks.”
The technology is exceedingly powerful for understanding gene regulation during different disease states, the researchers say. While previous techniques could only map a single type of protein at a time, ChIP–DIP can look at hundreds at once to give a comprehensive picture in rare cell types and in both healthy and disease tissue samples from patients.
“We can apply ChIP–DIP to understand the epigenetic signatures [how environmental factors influence gene expression] of diseases,” says postdoctoral scholar Andrew Perez (Ph.D. ’24), co-first author on the new study. “We only need one sample to map hundreds of proteins at once.”
“This has been a long-standing goal of the genomics field for decades,” Guttman says. “ChIP–DIP changes the paradigm of what’s possible.”
More information:
Andrew A. Perez et al, ChIP-DIP maps binding of hundreds of proteins to DNA simultaneously and identifies diverse gene regulatory elements, Nature Genetics (2024). DOI: 10.1038/s41588-024-02000-5
Provided by
California Institute of Technology
Citation:
New method maps hundreds of proteins in cell nuclei simultaneously (2024, December 18)