BGI Genomics, in collaboration with Southwest University, the State Key Laboratory of Silkworm Genome Biology, and other partners, has constructed a high-resolution pangenome dataset representing almost the entire genomic content in a silkworm.
This research paper, providing genetic insights into artificial selection (domestication and breeding) and ecological adaptation, was published on September 24 in Nature Communications.
Previously, due to the scarcity of wild silkworms and technical limitations of former studies, many trait-associated sites were missing. This is the first research ever to digitize silkworm gene pool and create a “digital silkworm”, greatly facilitating functional genomic research, promoting precise breeding, and thus enabling additional silk use cases.
The team deeply re-sequence 1,078 silkworms (B. mori, including 205 local strains, 194 improved varieties, and 632 genetic stocks and 47 wild silkworms, B. mandarina) and assemble long-read genomes on 545 of these samples, generating 55.57 T of genomic data.
This pangenome dataset contains the most comprehensive information on the genomes of domestic and wild silkworm, and is the largest long-read pangenome in the world for plants and animals to date. At the same time, in-depth studies on various genetic variation, population structure, artificial selection and ecological adaptations and economic traits of silkworm have been carried out, yielding fruitful results.
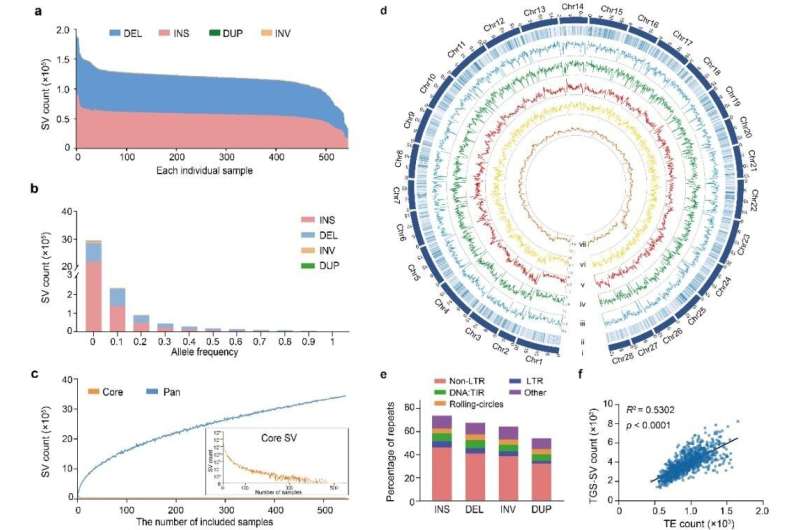
Characterization of structural variation in 545 silkworm genomes. © BGI Genomics
The origins of the domestic silkworm
The domestic silkworm, B. mori, domesticated from the wild mulberry silkworm, B. mandarina. It has an history of over 5,000 years, but its domestication origin location has long been an open question, due to a lack of strong biological evidence.
The material in this study represents the richest genetic diversity from all major sericulture regions of the world. The study found out that endemic species from China’s lower and middle Yellow River region are distributed at the base of the domestic silkworm branch on the evolutionary tree, thus suggesting that the domestic silkworm originated in this region. The available archaeological evidence, including a half cocoon excavated in 1926 at Xiyin Village, Xia County, Shanxi Province, and a stone-carved silkworm pupa excavated in 2019 at Shicun in the same county, provide important support to this conclusion.
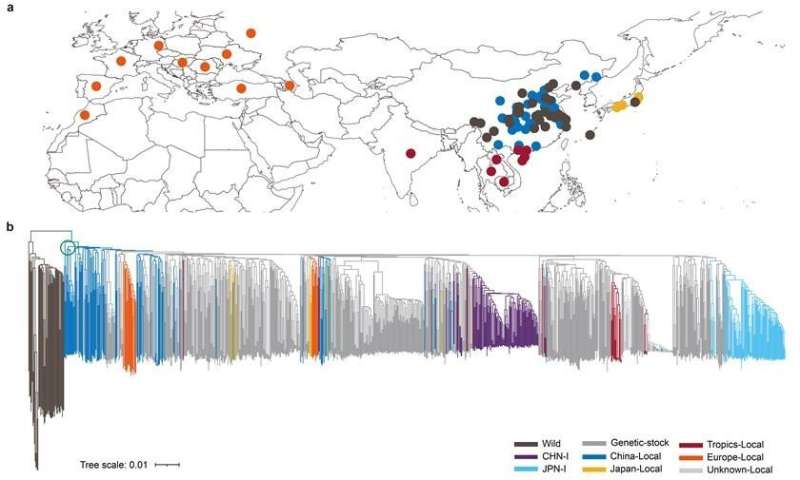
Geographic distribution and phylogenetic tree of silkworm. © BGI Genomics
Breaking the bottleneck in silkworm breeding
The traditional breeding of silkworms has a long and unique history, but since the 1990s remained stuck in a bottleneck. Systematic analysis of the genetic basis of domestication and improvement selection is essential to solve the unresolved issues in silkworm breeding. The team has identified 468 domestication-associated genes and 198 improvement-associated genes, of which respectively 264 and 185 are newly identified. These genes will be important candidate targets for molecular improvement of silkworm.
At the same time, it was found out that the Chinese and Japanese utility species share less than 3% of the improvement loci. This not only reveals the relatively independent breeding histories of Chinese and Japanese silkworm, but also explains why this shared genetic basis provides such hybrid advantages for both species. This result sheds new insights for future breeding of the silkworm.
Economic traits of silkworm breeding
Yield and quality of silk have long been targeted as the main economic criteria for artificial selection of silkworm. However, up to this date, little is known about how genes and loci control these quantitative traits. The pangenome is arguably the ‘nearest bridge’ between phenotypes, especially complex traits.
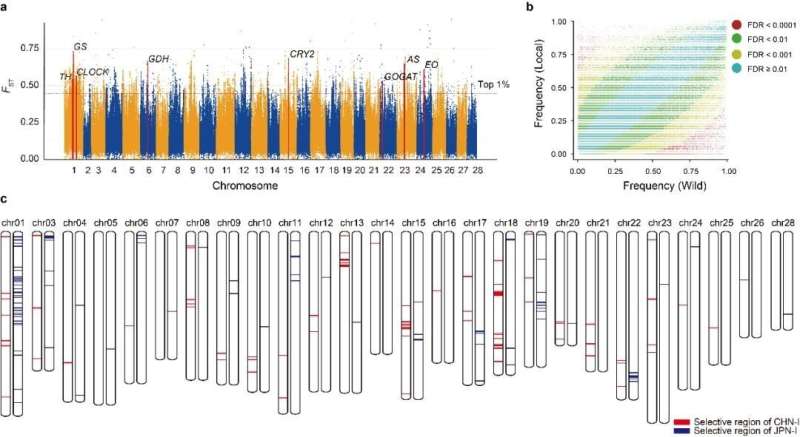
Silkworm domestication and breeding. © BGI Genomics
A case in point is the regulation of silk production by the cell cycle-related transcription factor BmE2F1, which was revealed through selection signaling and structural variation. CRISPR-cas9 mediated knockout of BmE2F1 reduces the number of silk gland cells by 7.68% and silk yield by 22%. Conversely, the transgenic overexpression of BmE2F1 increases the number of silk gland cells by 23% and silk yield by 16%.
Fine silk has unique applications and higher economic value, but the genetic basis of fiber fineness remained previously unknown. Analysis of rare variants in the genomes of slender varieties led to the identification of BmChit β-GlcNAcase, a gene controlling silk fineness that can significantly be detected in fine varieties, and CRISPR-cas9 mediated knockout, resulting in coarser silk fineness produced by domestic silkworms. This suggests this gene plays a key role in determining silk fineness.
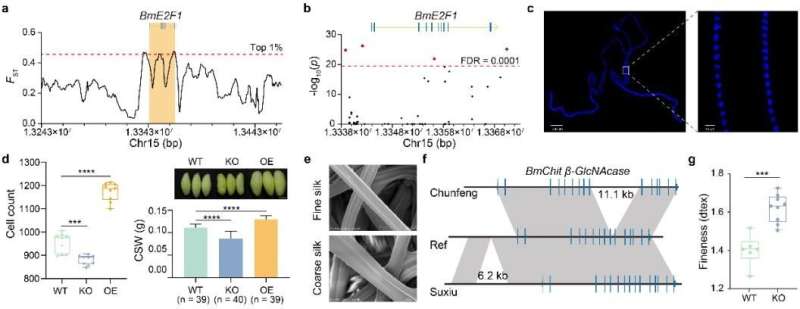
Genetic basis of silkworm economic traits. © BGI Genomics
Adaptive traits of silkworm breeding
Diapause is a common ecological adaptive trait in insects that ensures that insects can survive despite unfavorable environmental conditions. Although the diapause hormone was first identified in the silkworm in 1957, little information is available on the embryonic dipause gene. In this study, based on the analysis of the pnd strain and genomic structural variation in the silkworm, and functional validation by gene editing, the BmTret1-like gene revealed itself to be an important determinant of post-embryonic stalling. This is the first time that a post-embryonic determinant gene has been identified in an insect.
This study reveals the complete pan-genome of the silkworm to unlock artificial selection and ecological adaptation insights. Shuaishuai Tai, co-author and BGI Genomics senior researcher commented, “With comprehensive sampling and dataset combined with a variety of experiments to identify genes for future potential study, we hope to accelerate the process of silkworm molecular design breeding.”
More information:
Xiaoling Tong et al, High-resolution silkworm pan-genome provides genetic insights into artificial selection and ecological adaptation, Nature Communications (2022). DOI: 10.1038/s41467-022-33366-x
Provided by
BGI Genomics
Citation:
Scientists sequence the world’s largest pangenome to help unlock genetic mysteries behind finer silk (2022, September 30)



