Analytical chemistry aims to efficiently discriminate between two amino acids. In a new report in Science Advances, Zihao Liu and a research team in Chemistry, Physics, and Materials Science in China and Japan, used a single-molecule electrical method based on molecular nanocircuits to form stable graphene-molecule-graphene single-molecule junctions. The team developed these molecular junctions by covalently bonding a molecular machine. Then they used pH to vary the type and charge of the amino acids to find distinct multimodal current fluctuations originating from diverse host guest interactions relative to theoretical calculations. The conductance data produced characteristic leveling times and shuttling rates for each amino acid to allow accurate and real-time in situ measurements. The scientists tested four amino acids and their enantiomers (chiral molecules that are mirror images of one another) to show the ability to distinguish between them in a few microseconds to provide a simple and precise method for amino acid identification as well as single-molecule protein sequencing.
Building blocks of proteins
Amino acids form building blocks of proteins, molecules with special physiological functions and key structural elements in pharmaceuticals with applications in life sciences. Researchers can recognize enantiomers of amino acids to provide important information relative to chiral recognition and physiological functions in biological systems. Detecting the structure, enantiomeric purity and the dynamic behavior of amino acids can promote the production of new techniques for protein sequencing and pharmaceutical investigations. The detection and identification of amino acids with different structures and chirality are critically important across proteomics, pharmaceuticals and nanobiotechnology. However, existing molecular machines only function by comparing the thermodynamic parameters of the ensemble, which is insufficient. It is therefore important to develop a general molecular technique to revolutionize the existing methods and recognize target molecules with high accuracy. In this work, Liu et al. demonstrated a single-molecule technique to directly identify different proteinogenic amino acids and their enantiomers via accurate dynamic measurements of host-guest interactions based on a single-molecule technique known as graphene-molecule-graphene single-molecule junctions abbreviated as GMG-SMJs to covalently incorporate individual molecular systems behaving as the conductive channel into an electrical nanocircuit.
The new approach provided a robust platform to form single-molecule electronics to create molecular optoelectronic devices. To develop the constructs, Liu et al. covalently sandwiched a molecular machine between a pair of nano-gapped graphene point contacts for amino acid detection and chirality recognition. The outcomes will open new routes to develop the nanotechnology of accurate single-molecule protein sequencing for practical applications.
Device development, electrical characterization and real-time measurements
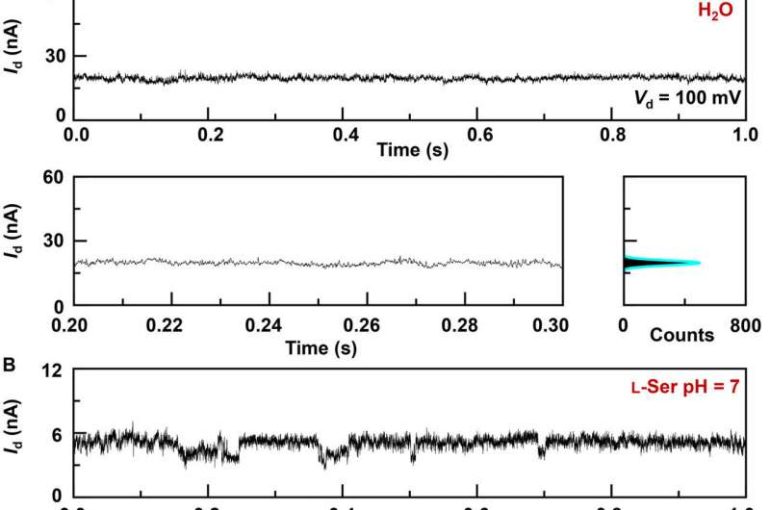
The research team synthesized single-layer graphene using chemical vapor deposition on copper foils and transferred it to silicon dioxide/silicon wafers and patterned the metal electrodes using photolithography. To develop the nano-gapped graphene point contact electrodes, they used a dash-line lithographic method. They then constructed GMG-SMJs (graphene-molecule-graphene single-molecule junctions) by covalently connecting a single molecular machine with graphene electrodes via amide linkages. The team measured the current-voltage curves of the devices at a variety of stages to identify the formation of GMG-SMJs. The charge transport through the junction resulted from single-molecule connections. They then conducted time-dependent electrical characterizations to monitor the conductance of permethylated-β-cyclodextrin (PM-β-CD) single molecular junctions (SMJs) in real-time. They first measured the constructs in water and then in diverse solutions of different amino acids. The representative current-time trajectories revealed multiple-peak distributions for different amino acids in solution such as L-serine and L-alanine. The multiple distinct states of each amino acid only originated from the association/dissociation process between the functional PM-β-CD center and the surrounding amino acids.
Theoretical and statistical analysis
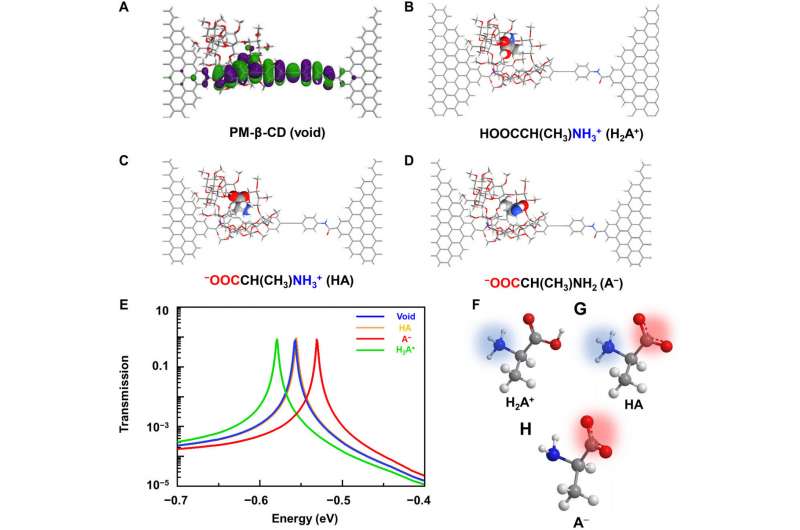
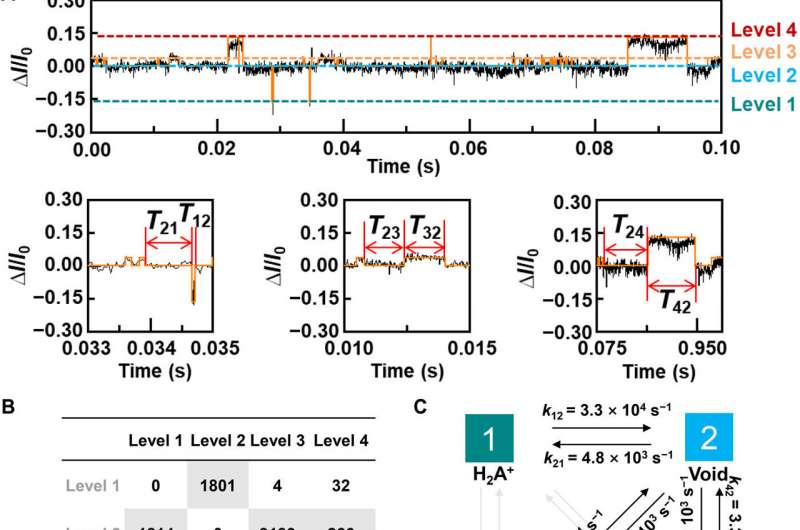
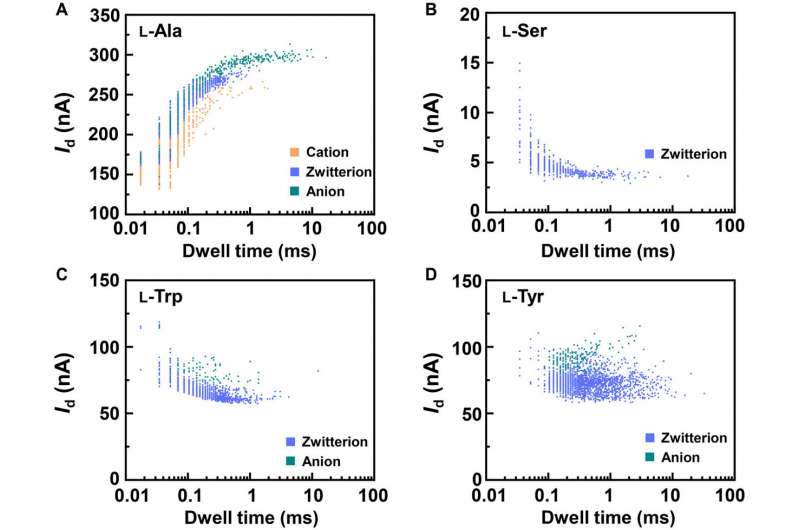
To better understand the correlation between host-guest interactions in the L-alanine based PM-β-CD single molecule junctions, Liu et al. calculated the transmission spectra of the PM-β-CD host with differently charged L-Alanine guests. To accomplish this, they used a nonequilibrium Green’s function technique based on density functional theory as implied in the Atomistix Toolkit package. They noted the conductance contribution from the perturbed highest occupied molecular orbital (p-HOMO) to be dominant at low bias voltages as reflected with transmission spectra. The transmission spectra of the configurations were significantly different near the Fermi level of electrodes to afford different conductance stages. The researchers then analyzed the transitions between each level during amino acid recognition. They used L-alanine as an example to observe reversible transitions from the dissociation to form cations, zwitterions, and anion forms. The dissociation level dominated the device conductance, and the team collected the information using a four-state model, which described the transitions to understand the capacity to detect different structures of amino acids when interacting with PM-β-CD. The amino acids had at least two association processes for the carboxylic group and the other amino group. The greater the number of states recorded, the more accurate the recognition of results were.
Enantiomer recognition and outlook
Due to the similarity between enantiomers, it was more challenging to differentiate the structures compared to species identification. The associated energy differences were small and close to computational errors. To overcome these complexities, the team established a distinct “fingerprint database” for each amino acid to compare the conductance and kinetic data of different enantiomers. The team then achieved the current change and relaxation time data and compared them with the fingerprint database. Due to this universality, the method can also recognize chiral functional drug molecules to establish a broad approach for biomolecular detection at the single molecule-level.
In this way, Zihao Liu and colleagues presented a practical single-molecule approach for real-time electrical recognition of amino acids with different structures and chirality within microseconds. The host-guest dynamic processes could be observed for diverse amino acids at the level of the anion, zwitterion and cation shuttling. Using the permethylated-β-cyclodextrin (PM-β-CD) complex, the team recognized differently charged states of amino acids based on current fluctuation range and thermodynamic/kinetic parameters. The technique can revolutionize existing methods to accurately sequence single-molecule gene/protein sequencing toward universal applications. The method can also offer a universal tool to recognize many important molecules in environmental or biological systems to understand the basis of life at the molecular level.
What is a protein? A biologist explains
More information:
1. Liu Z. et al. A single-molecule electrical approach for amino acid detection and chirality recognition, Science Advances, 10.1126/sciadv.abe4365
2. Sohrabi C. et al. Methods for generating and screening libraries of genetically encoded cyclic peptides in drug discovery. Nature Reviews Chemistry, doi.org/10.1038/s41570-019-0159-2
3. Oukhaled A. et al. Electrical recognition of the twenty proteinogenic amino acids using an aerolysin nanopore. Nature Biotechnology, ttps://doi.org/10.1038/s41587-019-0345-2
2021 Science X Network
Citation:
A single-molecule electrical approach for amino acid detection and chirality recognition (2021, March 19)
retrieved 20 March 2021
from https://phys.org/news/2021-03-single-molecule-electrical-approach-amino-acid.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.