Like Bill Murray in the movie “Groundhog Day,” bacteria species in a Wisconsin lake are in a kind of endless loop that they can’t seem to shake. Except in this case, it’s more like Groundhog Year.
According to a study in Nature Microbiology, researchers found that over the course of a year, most individual species of bacteria in Lake Mendota rapidly evolve, apparently in response to dramatically changing seasons.
Gene variants would rise and fall over generations, yet hundreds of separate species would return, almost fully, to near copies of what they had been genetically prior to a thousand or so generations of evolutionary pressures. (Individual microbes have lifespans of only a few days—not whole seasons—so the scientists’ work involved comparing bacterial genomes to examine changes in species over time.)
This same seasonal change played out year after year, as if evolution was a movie run back to the beginning each time and played over again, seemingly getting nowhere.
“I was surprised that such a large portion of the bacterial community was undergoing this type of change,” said Robin Rohwer, a postdoctoral researcher at The University of Texas at Austin in the lab of co-author Brett Baker. “I was hoping to observe just a couple of cool examples, but there were literally hundreds.”
Rohwer led the research, first as a doctoral student working with Trina McMahon at the University of Wisconsin-Madison and then at UT.
Lake Mendota changes greatly from season to season—during the winter, it’s covered in ice, and during the summer, it’s covered in algae. Within the same bacterial species, strains that are better adapted to one set of environmental conditions will outcompete other strains for a season, while other strains will get their chance to shine during different seasons.
The team used a one-of-a-kind archive of 471 water samples collected over 20 years from Lake Mendota by McMahon, Rohwer and other UW-Madison researchers as part of long-term monitoring projects.
For each water sample, they assembled a metagenome, all of the genetic sequences from fragments of DNA left behind by bacteria and other organisms. This resulted in the longest metagenome time series ever collected from a natural system.
“This study is a total game changer in our understanding of how microbial communities change over time,” Baker said. “This is just the beginning of what these data will tell us about microbial ecology and evolution in nature.”
This archive also revealed longer-lasting genetic changes.
In 2012, the lake experienced unusual conditions: the ice cover melted early, the summer was hotter and drier than usual, the flow of water from a river that feeds into the lake dwindled, and algae, which are an important source of organic nitrogen for bacteria, were more scarce than usual.
As Rohwer and the team discovered, many of the bacteria in the lake that year experienced a major shift in genes related to nitrogen metabolism, possibly due to the scarcity of algae.
“I thought, out of hundreds of bacteria, I might find one or two with a long-term shift,” Rohwer said. “But instead, one in five had big sequence changes that played out over years. We were only able to dig deep into one species, but some of those other species probably also had major gene changes.”
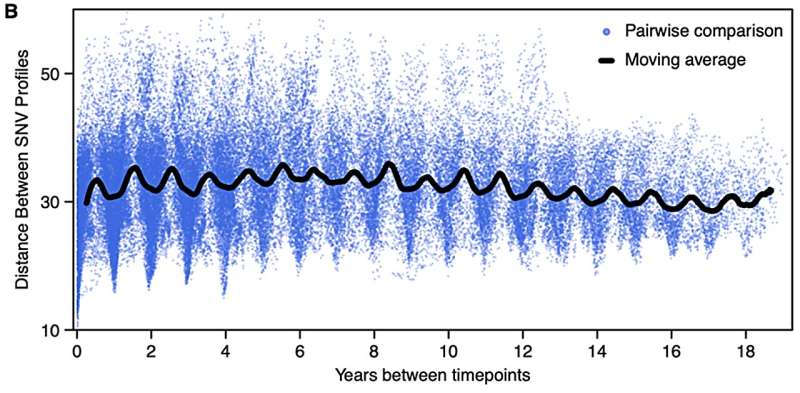
Most species of bacteria in Lake Mendota rapidly evolved with the change of seasons, returning to a similar state every year, for 20 years. The blue dots represent how much individual species within the genus Nanopelagicus changed genetically over time. The black line represents a 6-month moving average. © University of Texas at Austin
Climate scientists predict more extreme weather events—like the hot, dry summer experienced at Lake Mendota in 2012—for the midwestern U.S. during the coming years.
“Climate change is slowly shifting the seasons and average temperatures, but also causing more abrupt, extreme weather events,” Rohwer said. “We don’t know exactly how microbes will respond to climate change, but our study suggests they will evolve in response to both these gradual and abrupt changes.”
Discover the latest in science, tech, and space with over 100,000 subscribers who rely on Phys.org for daily insights.
Sign up for our free newsletter and get updates on breakthroughs,
innovations, and research that matter—daily or weekly.
Unlike another famous bacterial evolution experiment at UT, the Long-Term Evolution Experiment, Rohwer and Baker’s study involved bacterial evolution under complex and constantly changing conditions in nature. The researchers used the supercomputing resources at the Texas Advanced Computing Center (TACC) to reconstruct bacterial genomes from short sequences of DNA in the water samples.
The same work that took a couple of months to complete at TACC would have taken 34 years with a laptop computer, Rohwer estimated, involving over 30,000 genomes from about 2,800 different species.

The team analyzed genetic material from microbes in a one-of-a-kind archive of water samples collected over 20 years from Lake Mendota in Wisconsin. © Robin Rohwer/University of Texas at Austin.
“Imagine each species’ genome is a book, and each little DNA fragment is a sentence,” Rohwer said. “Each sample has hundreds of books, all cut up into these sentences. To reassemble each book, you have to figure out which book each sentence came from and put them back together in order.”
Other co-authors of the new study are Mark Kirkpatrick at UT; Sarahi Garcia of Carl von Ossietzky University of Oldenburg (Germany) and Stockholm University; and Matthew Kellom of the U.S. Department of Energy’s Joint Genome Institute.
This is one of two related papers published in the journal; the companion paper focuses on the ecology and evolution of viruses from the same lake samples.
More information:
Robin R. Rohwer et al, Two decades of bacterial ecology and evolution in a freshwater lake, Nature Microbiology (2025). DOI: 10.1038/s41564-024-01888-3
Zhichao Zhou et al, Unravelling viral ecology and evolution over 20 years in a freshwater lake, Nature Microbiology (2025). DOI: 10.1038/s41564-024-01876-7
Provided by
University of Texas at Austin
Citation:
Lake bacteria evolve like clockwork with the seasons, study reveals (2025, January 3)



