Researchers at Umeå University have discovered how a certain type of protein moves for DNA to be copied. The discovery could have implications for understanding how antibiotic resistance genes spread between bacteria.
“Studying DNA replication is a good starting point for potentially identifying targets for future drug development,” says Ignacio Mir-Sanchis, lead researcher in the group at Umeå University that published the study.
All cellular organisms must replicate their genetic material, DNA, to proliferate, so that one copy goes to a daughter cell and the other copy goes to the other daughter cell. The DNA molecule can be likened to a very long string of beads, where the beads are the building blocks or units.
The string of pearls has two strands that are intertwined to form a spiral structure, a double helix. To duplicate its genetic material, the cell must go from one to two DNA molecules, a process called DNA replication, and it starts by separating the two strands of DNA. To separate the two strands, cells have specialized proteins called helicases.
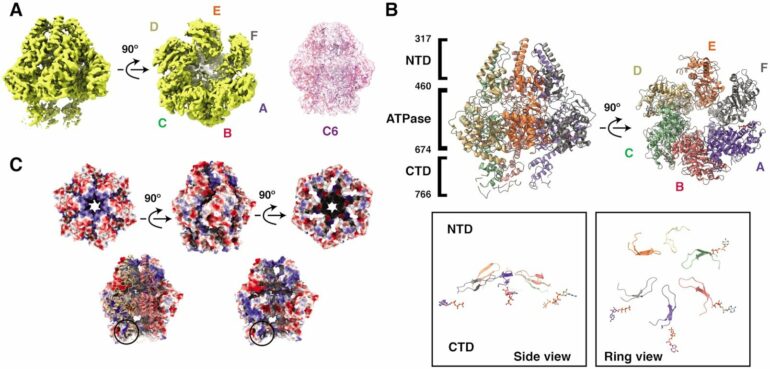
A research group at the Department of Medical Biochemistry and Biophysics at Umeå University has found how helicases interact and move on DNA to separate its strands. The discovery was made possible by so-called cryo-electron microscopy, for which Umeå has one of Sweden’s most advanced facilities. This technique allows scientists to take snapshots of a single molecule. By combining millions of snapshots, they can make a movie and see how the helicases move.
“When we analyzed our snapshots, we saw that the helicases move different parts, called domains, via two separate motions. Two domains rotate and tilt towards each other. These movements give us clues about how these helicases move on DNA and separate the two strands,” says Cuncun Qiao, a postdoctoral researcher in the team and first author of the paper.
Mir-Sanchi’s lab focuses on infection biology and studies the Staphylococcus aureus bacterium. The researchers are interested in understanding the DNA replication of S. aureus, of viruses that infect S. aureus (called bacteriophages) and of viral satellites. Viral satellites are viruses that parasitize other viruses.
S. aureus infects and kills millions of people worldwide and is considered a major threat because the bacterium has become resistant to almost all antibiotics. Interestingly, the genes involved in antibiotic resistance are sometimes also present in viral satellites, making the work even more medically relevant.
“The findings broaden our understanding of how antibiotic resistance genes spread, although it is worth noting that the movements we have identified here have also been seen in helicases found in eukaryotic viruses and even in human cells. It’s always surprising how important mechanisms are conserved from bacteriophages to humans,” says Ignacio Mir-Sanchis.
The findings are published in the journal Nucleic Acids Research.
More information:
Cuncun Qiao et al, Staphylococcal self-loading helicases couple the staircase mechanism with inter domain high flexibility, Nucleic Acids Research (2022). DOI: 10.1093/nar/gkac625
Citation:
Movements in proteins reveal information about antibiotic resistance spreading (2023, January 27)