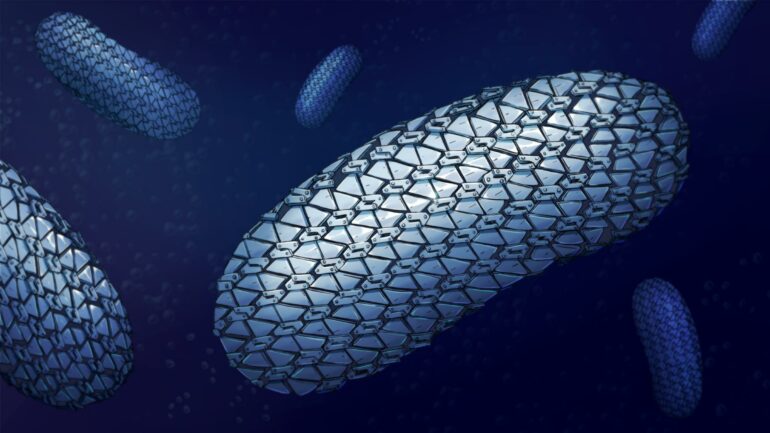
The structure of the protective armor of superbug C. difficile has been revealed for the first time, showing the close-knit yet flexible outer layer, which resembles chainmail. This assembly prevents molecules getting in and provides a new target for future treatments, according to the scientists who have uncovered it.
Dr. Paula Salgado, senior lecturer in macromolecular crystallography, who led the research at Newcastle University said, “This opens the possibility of developing drugs that target the interactions that make up the chainmail. If we break these, we can create holes that allow drugs and immune system molecules to enter the cell and kill it.”
One of the current challenges in the fight against infections is the growing resistance of bacteria to antibiotics. Antimicrobial resistance (AMR), was declared by WHO as one of the top 10 global public health threats facing humanity.
Publishing in Nature Communications, the team of scientists from Newcastle, Sheffield and Glasgow Universities, together with colleagues from Imperial College and Diamond Light Source, outline the structure of the main protein, SlpA, that forms the links of the chainmail and how they are arranged to form a pattern and create this flexible armor.
“I started working on this structure more than 10 years ago, it’s been a long, hard journey but we got some really exciting results. Surprisingly, we found that the protein forming the outer layer, SlpA, packs very tightly, with very narrow openings that allow very few molecules to enter the cells. S-layer from other bacteria studied so far tend to have wider gaps, allowing bigger molecules to penetrate. This may explain the success of C. diff at defending itself,” said Salgado

PhD student Paola Lanzoni-Mangutchi and Dr Anna Barwinska-Sendra from Newcastle University collecting data on I24 beamline at Diamond Light Source. © Newcastle University
One of the many ways that diarrhea-causing superbug Clostridioides difficile has to protect itself from antibiotics is a special layer that covers the cell of the whole bacteria—the S-layer. This flexible armor protects against the entry of drugs or molecules released by our immune system to fight bacteria.
The team determined the structure of the proteins and how they are arranged using a combination of X-ray and electron crystallography. Making a natural 2D array crystallize as a 3D crystal was not easy, and the crystals had many problems.
“Even when crystals were obtained, not all diffracted well, so Diamond synchrotron was essential for the success of the project. The work relied particularly on I24, the microfocus beamline to test many hundreds of crystals and screen for the best spot in the best crystal to collect the best data set. Getting native data was not the end of the story as there were no models to allow structure determination. Staff across the MX beamlines at Diamond were always eager to help solve this problem and willing to try new approaches and accommodate requests for specialized beamtime. But the crucial experiment was using the unique, long-wavelength I23 beamline, which allowed using the native sulfur atoms in SlpA as sources of information required to start building a model,” said Salgado.
The data collected on I23 allowed Senior Beamline Scientist Dr. Kamel El Omari to find the positions of the sulfur atoms and generate an initial partial model. This was the starting point that, together with higher resolution data collected at I24 and I04 beamlines at Diamond, allowed Dr. Salgado and her team to build the full SlpA structure.
Dr. El Omari said: “It was a pleasure to be part of this long standing and exciting project. It is a nice example showing how collaborations and access to state-of-the-art facilities like Diamond Light Source are successfully supporting the scientific community.”
So-called superbugs have multiple ways to resist antibiotics and can combine multiple resistance mechanisms. C. diff, is a superbug that infects the human gut and is resistant to all but three current drugs. Not only that, but it also becomes a problem when antibiotics are taken, as the good bacteria in the gut are killed alongside those causing an infection. As C. diff is resistant, it can grow and result in ill effects, ranging from diarrhea to death, due to massive lesions in the gut. Another problem is the fact that the only way to treat C.diff is to take more antibiotics, so the cycle is restarted and many people get repeat infections.
Determining the structure allows the possibility of designing C. diff-specific drugs to break the S-layer, the chainmail, and fight infections.
From Dr. Salgado’s team at Newcastle University, Ph.D. student Paola Lanzoni-Mangutchi and Dr. Anna Barwinska-Sendra unraveled the structural and functional details of the building blocks and determined the overall X-ray crystal structure of SlpA. Paola said: “This has been a challenging project and we spent many hours together, culturing the difficult bug and collecting X-ray data at the Diamond Light Source synchrotron.”
Dr. Barwinska-Sendra added: “Working together was key to our success, it is very exciting to be part of this team and to be able to finally share our work.”
Dr. Rob Fagan and Professor Per Bullough’s teams at the University of Sheffield carried out the electron crystallography work. Dr. Fagan said: “We’re now looking at how our findings could be used to find new ways to treat C. diff infections such as using bacteriophages to attach to and kill C. diff cells—a promising potential alternative to traditional antibiotic drugs.”
More information:
Paola Lanzoni-Mangutchi et al, Structure and assembly of the S-layer in C. difficile, Nature Communications (2022). DOI: 10.1038/s41467-022-28196-w
Provided by
Diamond Light Source
Citation:
Protective armor of superbug C. difficile revealed for the first time, providing target for treatments (2022, February 25)