Scientists from Skoltech and research centers in Sweden and Switzerland have presented results that explain the mechanism of interaction between bacteria and phages—viruses that infect bacterial cells. The discovery is an important step on the way to developing new ways to fight infections. The study is published in the journalCell Reports.
Disease-causing microorganisms become more resistant to antibiotics. Researchers across the world are searching for new mechanisms to combat pathogenic bacteria—one of them is phage therapy. Bacteriophages are natural “predators” and enemies of bacteria with a high infection specificity. They are harmless to the human organism, have a natural origin, and can be found wherever there is life and bacteria. Phages had been studied more than 100 years ago, but when antibiotics were discovered, research interest declined.
“Our laboratory at Skoltech pursues research into the new systems of bacterial immunity. In recent years, bioinformatics has allowed predictions of a plethora of novel antivirus systems and a new field emerged—microbial immunology,” says Head of the Laboratory of Metagenome Analysis and a study co-author Artem Isaev.
“Molecular mechanisms for most of those systems have not been uncovered yet, so one of the ways to learn how microbial immunity works is to find out how viruses managed to overcome the defense. The interaction between viruses and bacteria can be described as an arms race: when bacteria acquire a new defense strategy, it exerts great pressure on the opposing side—phages. In turn, phages that somehow acquire resistance to the bacterial immunity should emerge.
“It initiates a new stage of bacterial adaptation, and such intense competition leads to a wide variety of antiviral systems.”
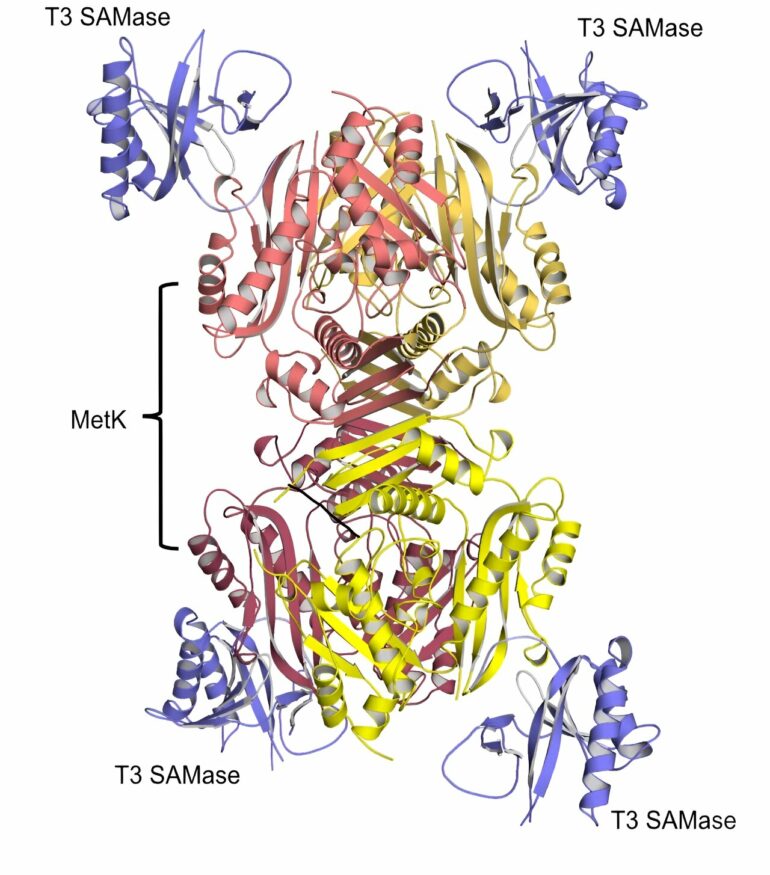
The authors studied the BREX (short for BacteRiophage EXclusion) immunity system, a protective mechanism of bacteria against phages, and showed that bacteriophage T3 can infect the BREX culture only when the special enzyme is expressed—the SAM lyase, which destroys S-adenosyl-methionine, an important cell metabolite.
“SAM is used as a donor of methyl groups, which can regulate the expression of genes. In the case of the BREX system, a methyl group also serves as a label, which allows bacteria to distinguish between their own DNA and the non-methylated DNA of the virus.
“Unexpectedly, the methylation function of BREX was not severely affected, while the BREX protection was completely disrupted by the SAMase expression. We can assume that SAM is also a cofactor necessary for inhibition of the virus infection by the BREX system,” says Artem Isaev.
According to the researchers, many countries are developing bacteriophage banks and support projects allowing the use of phages for therapeutic purposes. Modification of virus particles might allow the construction of modular bacteriophages effective against a specific pathogenic microorganism detected in the body of a particular patient.
“We still do not know much about the repertoire of anti-defense proteins encoded by phages, while these proteins are largely responsible for the success of bacterial cells’ infection in the natural environment, considering a huge variety of immune systems present in bacteria (including pathogenic species).
“Our research contributes to the establishment of the interaction network between viral anti-defense proteins and bacterial immunity systems. It has to be considered at the next stage of the phage therapy development when we will approach the creation of phages capable of killing specific pathogenic bacteria,” adds Artem Isaev.
More information:
Aleksandr Andriianov et al, Phage T3 overcomes the BREX defense through SAM cleavage and inhibition of SAM synthesis by SAM lyase, Cell Reports (2023). DOI: 10.1016/j.celrep.2023.112972
Provided by
Skolkovo Institute of Science and Technology
Citation:
Researchers uncover new ways to combat pathogenic bacteria (2023, August 18)