University of Maryland biologists developed the first mathematical simulations of bacterial communities that incorporate the complex interactions and rapid evolution among bacteria and reflect the tremendous species diversity seen in real life.
Their work, published January 4, 2022 in the Proceedings of the National Academy of Sciences, establishes a new theoretical framework for studying bacterial communities and lays the groundwork for improved probiotic and antibiotic therapies.
Studying bacterial communities is a bit like trying to understand wildlife in a world where elephants with wrinkly gray skin and elephants with feathers jostle for space at the watering hole next to birds with trunks, zebras with scales and lions that evolved a taste for grass. The next day, new animals may appear with even more crisscrossed traits.
In the bacterial world, thousands of species can exist side by side in a fluid state of evolution. Individual bacteria snatch up bits of DNA from their neighbors, rapidly acquiring new traits and blurring the lines between species. Adding to the mayhem, the community is in a constant state of war defined by complex and ever-shifting alliances. Some species gobble each other up or spew toxins to kill or incapacitate one another, while others seem to shield each other from aggressors.
“This type of very fluid state between species and these very complex interactions are not something we see in ecology and evolution at our human scale, so traditional theoretical models have not been effective for explaining how microbial communities assemble and maintain diversity,” said Anshuman Swain, a biological sciences Ph.D. student at UMD and lead author of the new study.
Having the proper tools to study microbial communities is important because microbes are essential to human health and life on Earth. Microbes cause and prevent disease, break down food and waste, and help regulate immune systems. And species diversity in bacterial communities is critical to everything from a healthy digestive system to a balanced response to antibiotics.
Conventional rules of biology suggest that competition between species should reduce diversity and lead to the rise of a few distinct, dominant species. Clearly, those rules don’t reflect the reality under the microscope. That’s partly because they generally only account for interactions between two species at a time and don’t consider the rapid speed of evolution seen in microbes.
The lack of an appropriately sophisticated theoretical framework that accounts for the complex rules driving community dynamics has hindered attempts to develop predictions that scientists can test with meaningful real-world experiments.
“There has been very little theory that incorporates complex interactions to tell the experimentalists what is possible, what to look for, or how to expect a community to behave under certain conditions,” Swain said. “It’s very difficult to do experiments with many species, all interacting and influencing interactions downstream in the community. And then how do you keep up with rapid mutations on top of that?”
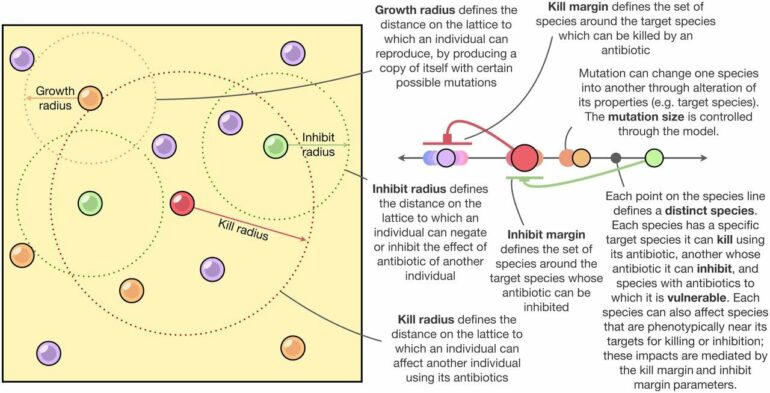
To address this problem, Swain worked with UMD’s Distinguished University Professor of Biology Bill Fagan and Levi Fussell of the University of Edinburgh to develop computer simulations that incorporated these unique complexities. After running over 10 million simulations with a variety of parameters, the team identified three key factors that influence species diversity, pointing for the first time to aspects of microbial community dynamics that experimentalists can focus on when asking questions about microbial diversity:
Higher-order interactions, which are interactions between two or more species that are regulated by one or more additional species. For example, species A produces a toxin that kills species B, but species C neutralizes that toxin if nearby and species B survives.Horizontal gene transfer, which creates a “continuous trait space.” This means genetic mixing causes a blending of traits between originally distinct species and leads to a community with a continuum of shared traits.High mutation rates among species in the community.
With a focus on these three factors, the researchers developed algorithms to simulate the growth and evolution of bacterial communities. Their models accounted for unprecedented diversity in communities with a nearly limitless number of species sharing traits along a continuum. (Traditional mathematical models that tried to incorporate complex interactions between species tended to be restricted to five or fewer distinct species.)
In their models, Swain and his colleagues found that the best predictor of species diversity was the time it took for a community to reach equilibrium, meaning that the number of bacteria and the physical space they occupied was relatively stable. When a community stabilized very rapidly or very slowly, diversity dropped and just a few species dominated.
But somewhere in between, diversity flourished. In that middle zone, the mutation rate and mobility of a bacterium in a community (how far it can spread into new territory) dictated the amount of diversity in a community.
Equipped with this new framework, experimentalists should be better prepared to address important questions, like how to develop antibiotics that maintain diversity in the gut microbiome or how to prevent antibiotic resistant strains of bacteria from dominating in an infection.
The research paper, “Higher-order effects, continuous species interactions, and trait evolution shape microbial spatial dynamics,” Anshuman Swain, Levi Fussell, and William F Fagan, was published in PNAS on January 4, 2022.
More information:
Anshuman Swain et al, Higher-order effects, continuous species interactions, and trait evolution shape microbial spatial dynamics, Proceedings of the National Academy of Sciences (2021). DOI: 10.1073/pnas.2020956119
Provided by
University of Maryland
Citation:
Rethinking the wild world of species diversity in microbes (2022, January 5)