Oak Ridge National Laboratory scientists had a problem mapping the genomes of bacteria to better understand the origins of their physical traits and improve their function for bioenergy production. The method they had successfully used for genetic mapping of plant feedstock for biofuels relied on creating and then analyzing diverse, sexually recombined variants, while bacteria generally reproduce asexually with limited diversity.
“Bacteria don’t reproduce the way we needed. They generally reproduce by division, where you go from one mother cell to two daughter cells that have essentially the same genome as the parent,” said Josh Michener, project lead in ORNL’s Biosciences Division.
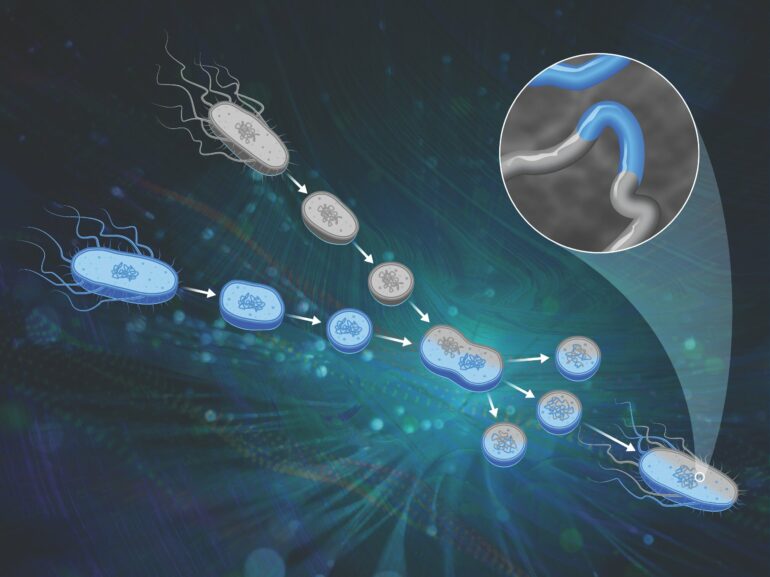
The solution came in a genetic research tool first developed in the 1970s: protoplast fusion. The scientists were able to cross Bacillus strains using this technique, and have produced a large population of genetically disparate organisms, which will result in greater insight into genomic variation and resulting physical characteristics.
Protoplast fusion is a classical genetic engineering method in which cells are stripped of their outer layer and chemically fused together, allowing recombination between the parents’ chromosomes. While researchers have used the technique for decades, first as a routine genetic technique and then as a tool to engineer traits like antibiotic production in microorganisms, the underlying chromosomal rearrangements were poorly understood.
“When scientists used this protoplast fusion technique in the past, they didn’t really know what the genomes of the progeny looked like due to the technical limitations of the day,” Michener said. “We were able to revive the technique and make it more straightforward. We basically benefited from 50 years of developments in the field so that we could do this kind of crossing and then examine dozens to hundreds of the progeny.”
The researchers used computational methods to analyze the genomes of the resulting progeny and map them back to their parents. “We could say, this little snippet of DNA in the genome came from parent A and then another little snippet from parent B, and so forth,” said Michener.
They found recombination throughout the genome across a variety of length scales, as outlined in Nucleic Acids Research. “We had both small and big pieces of DNA swapping, and the process looked to be mostly random. Those are exactly the characteristics that we wanted to enable genetic mapping.”
The team has since undertaken a total of four rounds of recombination, resulting in some 500 fully sequenced and highly shuffled progeny. A computational systems biology team led by ORNL’s Dan Jacobson is deploying machine learning methods to speed phenotyping and mapping across panels of bacteria to identify links between desired physical traits and their genetic underpinnings.
The result is a new microbial trait mapping capability that could hasten the design of microbes that are, for instance, better at breaking down plant biomass for the production of clean biofuels.
“One of the takeaways from the project is that there’s an awful lot of useful information in the old literature,” Michener said. “Sometimes it’s not all about the newest, snazziest approach.”
The discovery adds to the core strengths in genetic mapping that the Center for Bioenergy Innovation at ORNL is known for, Michener added. Other members of the research team were Delyana Vasileva, Jared Streich, Leah Burdick, Dawn Klingeman, Hari Chhetri, Christa Brelsford, Chris Ellis and Dan Close.
More information:
Delyana P Vasileva et al, Protoplast fusion in Bacillus species produces frequent, unbiased, genome-wide homologous recombination, Nucleic Acids Research (2022). DOI: 10.1093/nar/gkac025
Provided by
Oak Ridge National Laboratory
Citation:
Retro technique advances modern bacterial engineering for bioenergy (2022, April 6)