The vast majority of biological processes rely on proteins to carry out various jobs in all living cells, including neurons, which transmit information throughout the nervous system.
For Dr. Zachary Campbell, associate professor of biological sciences in the School of Natural Sciences and Mathematics at The University of Texas at Dallas, an overarching question is “How are proteins made in neurons?” The answer has been elusive.
Campbell is corresponding author of a study published Nov. 23 in Nature Communications that provides a significant step forward in delineating what controls the activation and deactivation of ribosomes, the microscopic factories inside cells that create proteins.
“Neurons do a remarkable number of things that are vital to life, including allowing us to sense the world around us,” said Campbell, who is also affiliated with UT Dallas’ Center for Advanced Pain Studies. “What memory, pain and fear all have in common is that they require protein synthesis. Thanks to revolutionary advances in microscopy, we’re getting our first atomic-scale glimpse at the protein-synthesis machinery obtained directly from tissues containing neurons.”
The process of making proteins begins when enzymes copy instructions from the DNA in a cell’s nucleus into messenger RNA, or mRNA. Strands of mRNA eventually arrive at ribosomes, where they are funneled through a channel and their instructions are read and translated into proteins.
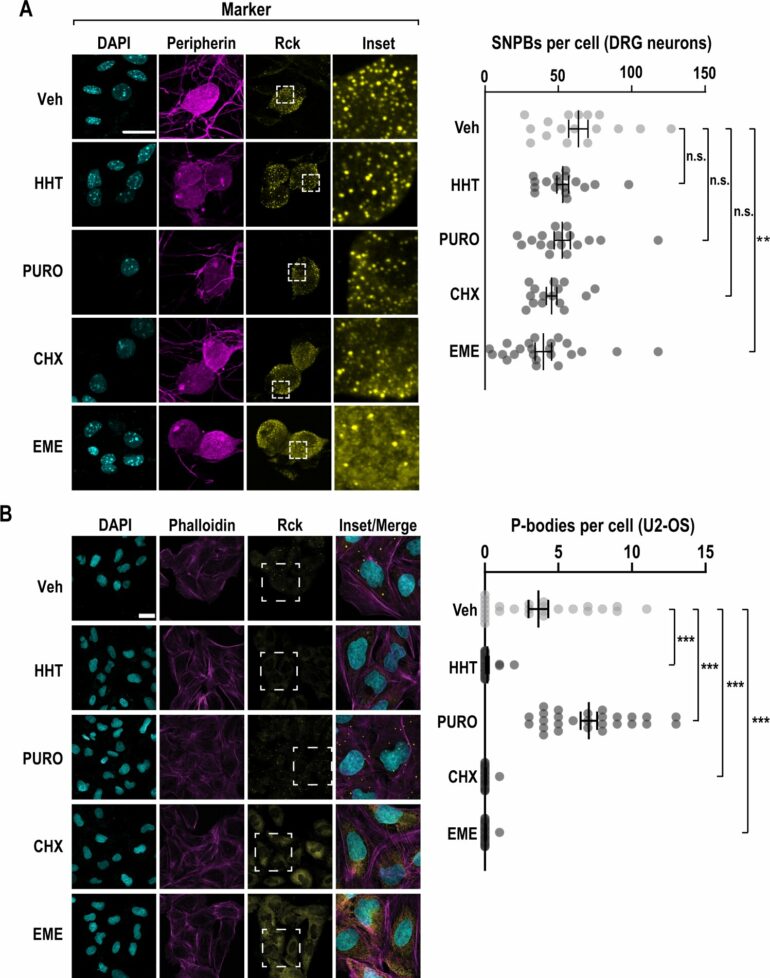
In many cell types including sensory neurons, however, some ribosomes are inactive. Just how a cell alternates the on/off switch for protein creation is poorly understood.
“Dysregulating this process can cause debilitating problems, which suggests that perhaps we could modulate the process in beneficial ways. Our research specifically sheds light on ribosome availability in sensory neurons,” Campbell said.
Within the cell, along with ribosomes, are processing bodies, or p-bodies—free-floating granules enriched in RNA degradation machinery and poorly translated mRNAs. Campbell described p-bodies as “mRNA purgatory,” where mRNA is neither made into protein nor actively degraded.
Campbell said that protein manufacturing machinery appears to be fundamental in sensitization of sensory neurons. While normally silent, harmful cues change their activity. This switching is fundamental to behaviors that promote injury avoidance and facilitate the healing process. Many aspects of this process remain a mystery, but the protein-making ribosomes are clearly integral. Ribosomes switch between active and inactive states, researchers found, because of a certain type of molecule called a kinase.
Molecular and cell biology graduate student Patrick Roark Smith BS’18, lead author of the paper, identified a kinase that promotes the preservation of inactive ribosomes and severely impedes the translation cycle.
“We’ve known for a very long time that inactive ribosomes exist, but we did not know how they are regulated,” Smith said. “We discovered an unexpected method of control: eEF2K (eukaryotic elongation factor-2 kinase) controls processing bodies as well as ribosomes. It seems to promote an inactive state of the ribosome where they can’t read the messenger RNA and make a protein.”
Campbell said the coordinate control of ribosomes and processing bodies by eEF2K appears to be specific to neurons, which is important because it implies a new way that protein synthesis is handled by the nervous system.
“This work really elevates our understanding of how eEF2K affects translation, and it’s in a completely different way than what we expected,” he said. “The kinase modifies a specific target, eEF2, which interacts with a partner protein, SERBP1, to block mRNA getting to the ribosome. The pair basically puts a cork in it.”
Campbell said this knowledge could lead to the development of drugs that manipulate this mechanism—causing ribosomes to become inactive—for therapeutic purposes.
“For neurons that transmit pain signals, if we can interfere with their ability to make the right proteins at the right time, perhaps we can fix disease states,” Campbell said.
The method behind this new information is single-particle cryo-electron microscopy (cryo-EM), an innovation that won its creators the 2017 Nobel Prize in chemistry.
“Cryo-EM gives us a much simpler way to get detailed structural maps and atomic-resolution views into molecules that were previously out of our reach,” Campbell said. “I remember thinking as a student that I’d be long dead before we knew what a ribosome looks like. Now, cryo-EM has shown us a multitude of states that ribosomes adopt. It’s kind of mind-blowing how advancements in methods transform fields.”
More information:
Patrick R. Smith et al, Functionally distinct roles for eEF2K in the control of ribosome availability and p-body abundance, Nature Communications (2021). DOI: 10.1038/s41467-021-27160-4
Provided by
University of Texas at Dallas
Citation:
Scientists shine new light on protein creation ‘on/off switch’ (2022, February 3)