All living organisms use proteins, which encompass a vast number of complex molecules. They perform a wide array of functions, from allowing plants to use solar energy for oxygen production to helping your immune system fight against pathogens to letting your muscles perform physical work. Many drugs are also based on proteins.
For many areas of biomedical research and drug development, however, there are no natural proteins that can serve as suitable starting points to build new proteins. Researchers designing new drugs to prevent COVID-19 infection, or developing proteins that can turn genes on or off or turn cells into computers, had to create new proteins from scratch.
This process of de novo protein design can be difficult to get right. Protein engineers like me have been trying to figure out ways to more efficiently and accurately design new proteins with the properties we need.
Luckily, a form of artificial intelligence called deep learning may provide an elegant way to create proteins that did not exist previously – hallucination.
New proteins created from scratch can be deployed to tackle a wide range of environmental and medical challenges.
Designing proteins from scratch
Proteins are made up of hundreds to thousands of smaller building blocks called amino acids. These amino acids are connected to one another in long chains that fold up to form a protein. The order in which these amino acids are connected to one another determines each protein’s unique structure and function.
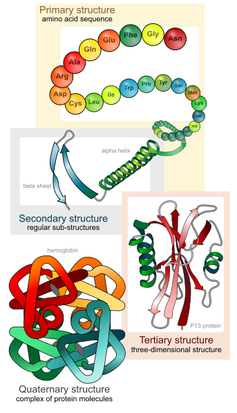
Proteins are composed of amino acid chains that fold into a protein.
LadyofHats/Wikimedia Commons
The biggest challenge protein engineers face when designing new proteins is coming up with a protein structure that will perform a desired function. To get around this problem, researchers typically create design templates based on naturally occurring proteins with a similar function. These templates have instructions on how to create the unique folds of each particular protein. However, because a template must be created for each individual fold, this strategy is time-consuming, labor-intensive and limited by what proteins are available in nature.
Over the past few years, various research groups, including the lab I work in, have developed a number of dedicated deep neural networks – computer programs that use multiple processing layers to “learn” from input data to make predictions about a desired output.
When the desired output is a new protein, millions of parameters describing different facets of a protein are put into the network. What’s predicted is a randomly chosen sequence of amino acids mapped onto the most probable 3D structure that sequence would take.
Network predictions for a random amino acid sequence are blurry, meaning the final structure of the protein is not very clear-cut, while both naturally occurring proteins…