Alternative splicing is a fundamental biological process that allows cells to make many different types of mRNAs and proteins from a limited number of genes. For many animals, including humans, it is a feature that is essential for the development of complex cells such as muscles or neurons.
Its fundamental importance means that alternative splicing is a very tightly regulated process. But a new study published today in the journal Science Advances has found evidence that the regulation of alternative splicing, which rarely goes wrong in healthy cells, goes haywire in an unexpected place—the cells of a newly formed embryo.
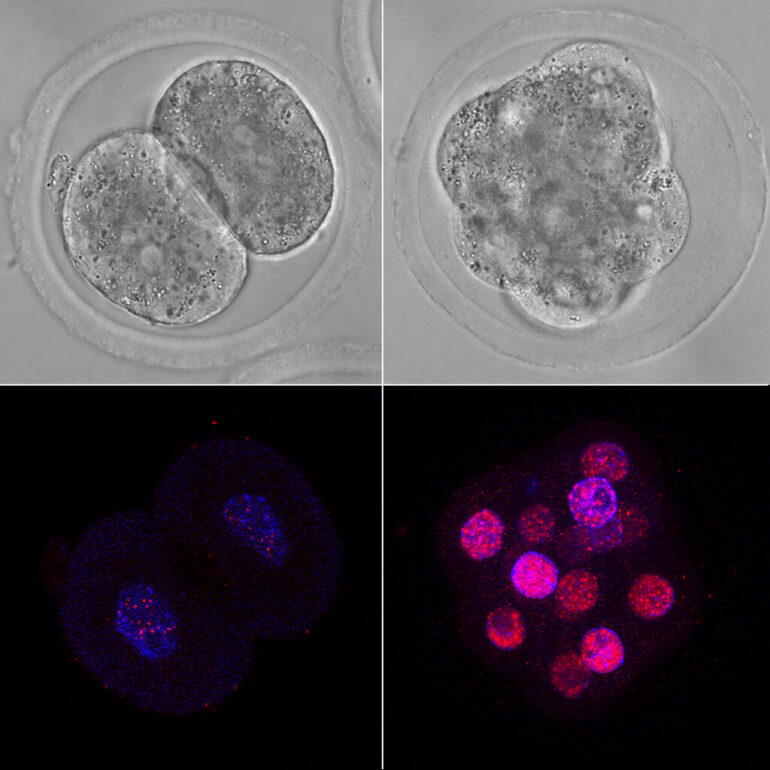
Researchers at the Centre for Genomic Regulation (CRG) in Barcelona made the discovery after creating an atlas of splicing events during the early development of cows, humans and mice.
They found that when human embryos are balls of just 8 cells, they express a huge variety of alternative mRNAs, so much so that the splicing diversity was the highest ever recorded across any cell or tissue studied to date. When the embryos transitioned to the next stage of development, their splicing activity returned to normal.
According to the authors of the study, this is evidence that the regulation of alternative splicing collapses temporarily at a crucial stage of development known as zygotic genome activation. This is when an early embryo transitions from using maternal resources such as proteins and RNA and making its own.
Importantly, the researchers believe the newly-discovered phenomenon occurs because it is developmentally programmed—a purposeful act of sabotage. “We think this happens because there are instructions in our genome that tell a few genes to not do their job at that developmental stage. The embryo cells mess up their splicing on purpose and they do so for a functional reason,” says ICREA Research Professor Manuel Irimia, senior author of the study.
An important clue for why the regulation of splicing fails at this crucial moment lies in the function of the proteins affected. The researchers found that splicing failure destroyed proteins responsible for responding to DNA damage.
“We saw that the DNA damage response at this stage of development was low. While splicing failure isn’t the only factor affecting this defense mechanism, it’s partly responsible for destroying the proteins involved. We don’t know why this happens, but it’s possibly because transcription itself carries a risk of DNA damage. As embryos activate their genome for the first time and start to transcribe, there may be trade-offs involved in order to avoid developmental failure,” says Dr. Barbara Pernaute, postdoctoral researcher at the CRG and co-first author of the study.
According to Dr. Pernaute, these results improve our understanding of how embryos develop during these early stages and could open doors for improvements in assisted reproductive technologies.
The findings could also be useful for advancing research efforts in the creation of totipotent cells from stem cells, a long-term aspiration for regenerative medicine. As these early embryonic cells are truly totipotent cells, knowledge of the mechanism could lead to advances that reverse engineer stem cells to induce totipotency.
“Recent studies carried out by other research groups around the world have shown that artificially inducing the mechanism we find in our study transforms stem cells into totipotent cells. We believe these programmed splicing failures also occur in other physiological contexts. We are only just scratching the surface for the importance this mechanism has for biological processes,” concludes Dr. Irimia.
More information:
Christopher D. R. Wyatt et al, A developmentally programmed splicing failure contributes to DNA damage response attenuation during mammalian zygotic genome activation, Science Advances (2022). DOI: 10.1126/sciadv.abn4935
Provided by
Center for Genomic Regulation
Citation:
Act of sabotage determines mammalian embryonic development (2022, April 13)