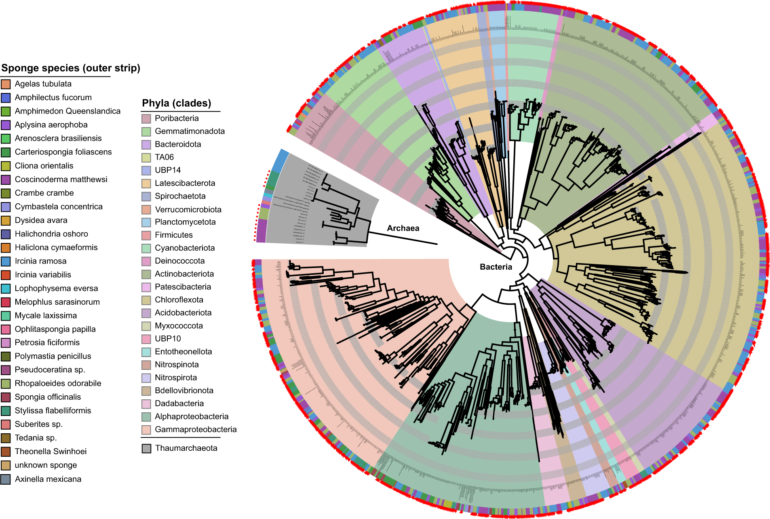
Sponges in coral reefs, less flashy than their coral neighbors but important to the overall health of reefs, are among the earliest animals on the planet. New research from UNH peers into coral reef ecosystems with a novel approach to understanding the complex evolution of sponges and the microbes that live in symbiosis with them. With this “genomic time machine,” researchers can predict aspects of reef and ocean ecosystems through hundreds of millions of years of dramatic evolutionary change.
“This study shows how microbiomes have evolved in a group of organisms over 700 million years old,” says Sabrina Pankey, a postdoctoral researcher at UNH and lead author of the study, published recently in the journal Nature Ecology & Evolution. “Sponges are increasing in abundance on reefs in response to climate change and they play an enormous role in water quality and nutrient fixation.”
The significance of the work transcends sponges, though, providing a new approach to understanding the past based on genomics. “If we can reconstruct the evolutionary history of complex microbial communities like this we can say a lot about the Earth’s past,” says study co-author David Plachetzki, associate professor of molecular, cellular and biomedical sciences at UNH. “Research like this could reveal aspects of the chemical composition of the Earth’s oceans going back to before modern coral reefs even existed, or it could provide insights on the tumult that marine ecosystems experienced in the aftermath of the greatest extinction in history that took place about 252 million years ago.”
The researchers characterized almost 100 sponge species from across the Caribbean using a machine-learning method to model the identity and abundance of every member of the sponges’ unique microbiomes, the community of microbes and bacteria that live within them in symbiosis. They found two distinct microbiome compositions that led to different strategies sponges used for feeding (sponges capture nutrients by pumping water through their bodies) and protecting themselves against predators—even among species that grew side by side on a reef.
“The types of symbiotic communities we describe in this paper are very complex, yet we can show they evolved independently multiple times,” says Plachetzki.
And, adds Pankey, “there’s something very specific about what these microbial communities are doing … sponges dozens of times have decided that this diverse arrangement of microbes works for them.”
Leveraging this new genomic approach, the researchers found that the origin of one of these distinct microbiomes, which had a high microbial abundance (HMA) of more than a billion microbes per gram of tissue, occurred at a time when the Earth’s oceans underwent a significant change in biogeochemistry coincident with the origins of modern coral reefs.
While machine learning and genomic sequencing generated the findings Plachetzki calls “a tour de force of microbial barcode sequencing,” this research began far from the lab, in the warm waters of the Caribbean.
“We dove for all 1,400 of these samples,” says Pankey, who went on five expeditions in 2017 and 2018 to collect sponges. “It was a monstrous collection,” she adds, acknowledging that SCUBA diving in the Caribbean has its rewards. The duo credits co-author Michael Lesser, UNH research professor emeritus, for establishing field work techniques, and their co-authors from the University of Mississippi and the Universidad Nacional del Comahue in Argentina for assisting with sponge collection and molecular identification. Former graduate student Keir Macartney also contributed to the study.
More information:
M. Sabrina Pankey et al, Cophylogeny and convergence shape holobiont evolution in sponge–microbe symbioses, Nature Ecology & Evolution (2022). DOI: 10.1038/s41559-022-01712-3
Provided by
University of New Hampshire
Citation:
Genomic time machine: From sponge microbiome, insights into evolutionary past (2022, April 14)