A team of researchers from the Eichman lab and associated with the Evolutionary Studies Initiative led a project that was recently published in mBio. Graduate student Noah Bradley and undergraduate student Katie Wahl (BA21, BCB) were co-first authors on the work studying chemical compounds produced by bacteria.
Specifically, the group was interested in a suite of chemicals known as natural products. These chemicals are produced by the organism for a specific purpose, but are often valuable because they may be used as antibiotics, anticancer agents, or other therapeutics. As new drug-resistant strains of diseases evolve, the importance of finding new weapons against resistant-diseases is ever increasing.
In this study, the researchers relied on a technique known as genome mining which Bradley describes as a useful tool in identifying gene clusters and natural products they produce. Historically, genome mining targets a specific mechanism used to produce a natural product. However, several mechanisms may produce similar natural products; thus, the team’s approach instead targets processes that provide bacteria with self-resistance towards a specific type of compound. Specifically, they were interested in mining for gene clusters that produce genotoxic natural products—those that can form a chemical bond to DNA.
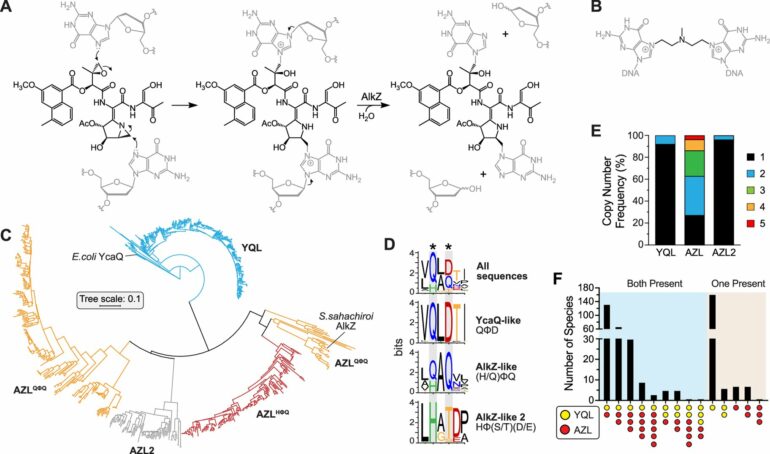
The group paid special attention to two bacterial DNA repair enzymes known as DNA glycosylases.
One, AlkZ, is found in Streptomyces sahachiroi while the other, YcaQ, is found in Escherichia coli. These enzymes—and those closely related—can remove DNA damage caused by genotoxins. However, the genomic environments and functions of AlkZ and YcaQ are very different.
AlkZ-like (AZL) enzymes tend to localize within biosynthetic gene clusters and according to Bradley, “AZL proteins seem to be tailored for self-resistance to specific natural products.”
However, YcaQ-like (YQL) enzymes have not been found in clusters and according to Wahl, “YQL proteins might be general caretakers to remove a variety of DNA damage.”
The pair of co-first authors are excited about what the results mean moving forward.
“In the short term, we hope that the approximately 70 uncharacterized gene clusters we identified in our study can be screened for the targeted discovery of novel or useful genotoxic natural products,” said Bradley.
“In the long term, we are hopeful that our self-resistance-guided genome mining framework will be applied in the scientific community for the targeted discovery of natural products of therapeutic benefit. We also envision the discovery of new DNA repair mechanisms through this approach,” added Wahl.
This project was borne out of creativity due to the COVID-19 pandemic forcing researchers out of the lab. Eichman reflected on the strong work ethic and positive attitudes of Bradley and Wahl.
“I enjoyed watching Noah and Katie execute a project that they designed specifically to be done while at home during the lab shut-down period. Not only did it help us all to stay engaged while being isolated, but this study has paved the way for an entirely new avenue of research for the lab,” he said.
This work was also a great example of the interdisciplinary research going on at Vanderbilt.
According to Bradley, “starting at the beginning of the pandemic, this project evolved into a fruitful collaboration with graduate student Jacob Steenwyk of the Rokas lab where they assisted on the phylogenetics and genome mining analysis.”
Bradley continued, “the Mass Spectrometry Research Center (MSRC) here at Vanderbilt also provided assistance with critical experiments in this project.”
More information:
Noah P. Bradley et al, Resistance-Guided Mining of Bacterial Genotoxins Defines a Family of DNA Glycosylases, mBio (2022). DOI: 10.1128/mbio.03297-21
Provided by
Vanderbilt University
Citation:
Researchers use new method to target potentially undiscovered beneficial therapeutic chemicals (2022, April 5)