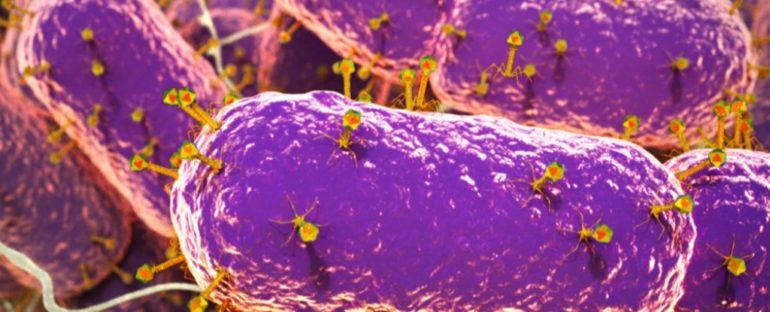
The coronavirus pandemic has had the world fixated on viruses like no time in living memory, but new evidence reveals humans never even notice the vast extent of viral existence – even when it’s inside us.
A new database project compiled by scientists has identified over 140,000 viral species that dwell in the human gut – a giant catalogue that’s all the more stunning given over half of these viruses were previously unknown to science.
If tens of thousands of newly discovered viruses sounds like an alarming development, that’s completely understandable. But we shouldn’t misinterpret what these viruses within us actually represent, researchers say.
“It’s important to remember that not all viruses are harmful, but represent an integral component of the gut ecosystem,” explains biochemist Alexandre Almeida from the European Molecular Biology Laboratory’s Bioinformatics Institute (EMBL-EBI) and the Wellcome Sanger Institute.
“These samples came mainly from healthy individuals who didn’t share any specific diseases.”
The new virus catalogue – called the Gut Phage Database (GPD) – was complied by analyzing over 28,000 individual metagenomes – publicly available records of DNA-sequencing of gut microbiome samples collected from 28 countries – along with almost 2,900 reference genomes of cultured gut bacteria.
The results revealed 142,809 viral species that reside in the human gut, constituting a specific kind of virus known as a bacteriophage, which infects bacteria, in addition to single-celled organisms called archaea.
In the mysterious environment of the gut microbiome – inhabited by a diverse mixture of microscopic organisms, encompassing both bacteria and viruses – bacteriophages are thought to play an important role, regulating both bacteria and the health of the human gut itself.
“Bacteriophages … profoundly influence microbial communities by functioning as vectors of horizontal gene transfer, encoding accessory functions of benefit to host bacterial species, and promoting dynamic co-evolutionary interactions,” the researchers write in their new paper.
For a long time, our knowledge of this phenomenon was stalled by limitations in our understanding of bacteriophage species.
In recent years, new advancements in metagenomic analyses have significantly expanded our awareness of the viral variety we’re looking at here – and perhaps none more so than the Gut Phage Database, which the researchers describe as a “massive expansion of human gut bacteriophage diversity”.
“To our knowledge, this set represents the most comprehensive and complete collection of human gut phage genomes to date,” the study authors write.
“Having a comprehensive database of high-quality phage genomes paves the way for a multitude of analyses of the human gut virome at a greatly improved resolution, enabling the association of specific viral clades with distinct microbiome phenotypes.”
Already, the database is updating what we know about viral behavior.
The research shows over one-third (36 percent) of viral clusters identified are not restricted to infecting a single species of bacteria, which means they can create gene flow networks across phylogenetically distinct bacterial species.
In addition, the researchers found 280 globally distributed viral clusters, including one newly identified clade, called Gubaphage, which appears to be the second most prevalent virus clade in the human gut, following what is known as the crAssphage group.
Given certain similarities between the two, the researchers initially thought the Gubaphage might belong to a proposed family of crAssphage-like viruses, before determining the clades were, in fact, distinct.
There is still so much to learn, and not just on the Gubaphage – but about more multitudes of viruses than we ever dared to dream. Thanks to research efforts like this, though, tomorrow’s discoveries are closer, and new insights will come faster.
“Bacteriophage research is currently experiencing a renaissance,” says microbiologist Trevor Lawley from the Wellcome Sanger Institute.
“This high-quality, large-scale catalogue of human gut viruses comes at the right time to serve as a blueprint to guide ecological and evolutionary analysis in future virome studies.”
The findings are reported in Cell.