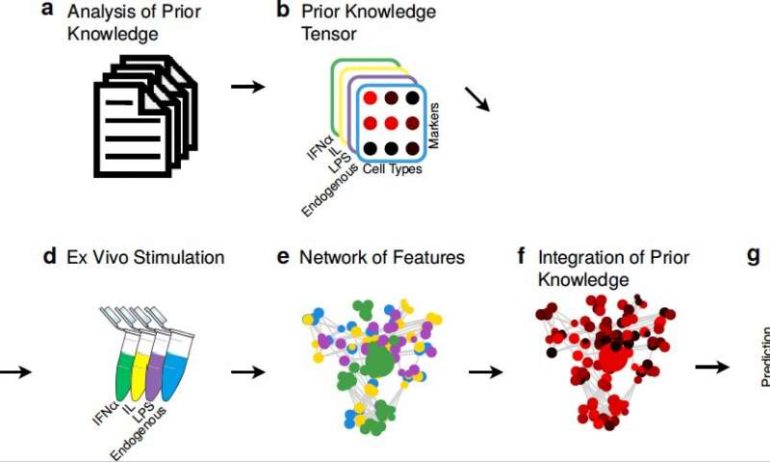
The complex network of interconnected cellular signals produced in response to changes in the human body offers a vast amount of interesting and valuable insight that could inform the development of more effective medical treatments. In peripheral immune cells, these signals can be observed and quantified using a number of tools, including cell profiling techniques.
Single-cell profiling techniques such as polychromatic flow and mass cytometry have improved significantly over the past few years and they could now theoretically be used to obtain detailed immune profiles of patients presenting a number of symptoms. Nonetheless, the limited sample sizes of past studies and the high dimensionality of the patient data collected so far increase the chances of false-positive discoveries, which in turn lead to unreliable immune profiles.
Conducting studies on larger groups of patients could improve the effectiveness of these cell-profiling techniques, allowing medical researchers to gain a better understanding of the patterns associated with medical conditions. Gathering data from many patients, however, can be both expensive and time consuming.
Researchers at Stanford University School of Medicine have recently developed immunological Elastic-Net (iEN), a machine-learning model that predicts cellular responses based on mechanistic immunological knowledge. In a paper published in Nature Machine Intelligence, they demonstrated that incorporating this immunological knowledge into their model’s prediction processes increased its predictive power on both small and large patient datasets.
“Our methodology allows us to leverage previous studies to increase our models’ accuracy without enrolling additional patients,” Nima Aghaeepour, one of the researchers who led the study, together with Anthony Culos, Martin Angst, and Brice Gaudilliere, told TechXplore. “A key advantage of our method is that it does not limit the data-driven nature of the models. In cases where the collected data disagrees with prior knowledge, our algorithm is allowed to reduce the importance of prior knowledge and instead focus on raw data if that proves to be the stronger solution.”
In scenarios where medical researchers must consider a wide number of dimensions, various features can be equally valuable for making predictions. Therefore, instead of discarding variables that are not consistent with prior immunological data, the machine learning algorithm developed by Aghaeepour and his colleagues selects all immune features that it finds to have a strong predictive value and relevance.
So far, the researchers have evaluated the performance of their machine-learning algorithm in three independent studies. In all of these studies, they found that their model could predict clinically relevant outcomes based on both simulated data and mass cytometry data generated from the blood of patients.
“In our paper, we include two real-world clinical examples in which the iEN pipeline increased our accuracy for modeling of pregnancy and periodontal disease,” Aghaeepour said. “We have several other exciting use cases that we cannot wait to see published, including recovery from surgery, Alzheimer’s disease, and Parkinson’s disease.”
In the future, the machine learning platform developed by Aghaeepour and his colleagues could aid the study of numerous diseases, medical conditions and neurological disorders. The data used by the researchers and the iEN algorithm are available online, so they could soon be accessed and used by other research teams worldwide.
“We are now also working toward developing versions of the algorithm that are applicable to other types of biological datasets,” Aghaeepour said. “A primary example of this are multiomics studies in which several omics technologies are used simultaneously for profiling of the immune system. We believe that these datasets provide unique opportunities for encoding prior knowledge into machine learning algorithms.”
New algorithm predicts likelihood of acute kidney injury
More information:
Integration of mechanistic immunological knowledge into a machine learning pipeline improves predictions. Nature Machine Intelligence(2020). DOI: 10.1038/s42256-020-00232-8.
2020 Science X Network
Citation:
A machine leaning model that incorporates immunological knowledge (2020, October 28)
retrieved 28 October 2020
from https://techxplore.com/news/2020-10-machine-incorporates-immunological-knowledge.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.