The Research Brief is a short take about interesting academic work.
The big idea
Diagram of the biological classification system, showing phylum is a broad grouping.
VectorMine/iStock via Getty Images Plus
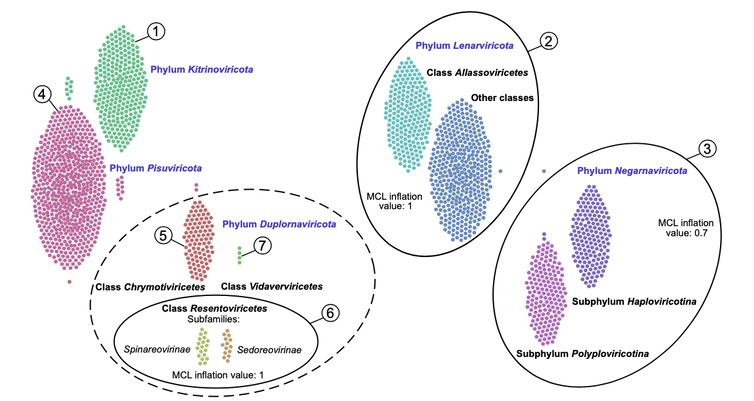
An analysis of the genetic material in the ocean has identified thousands of previously unknown RNA viruses and doubled the number of phyla, or biological groups, of viruses thought to exist, according to a new study our team of researchers has published in the journal Science.
RNA viruses are best known for the diseases they cause in people, ranging from the common cold to COVID-19. They also infect plants and animals important to people.
These viruses carry their genetic information in RNA, rather than DNA. RNA viruses evolve at much quicker rates than DNA viruses do. While scientists have cataloged hundreds of thousands of DNA viruses in their natural ecosystems, RNA viruses have been relatively unstudied.

There are more RNA viruses in the oceans than researchers previously thought.
Guillermo Domínguez Huerta, CC BY-ND
Unlike humans and other organisms composed of cells, however, viruses lack unique short stretches of DNA that could act as what researchers call a genetic bar code. Without this bar code, trying to distinguish different species of virus in the wild can be challenging.
To get around this limitation, we decided to identify the gene that codes for a particular protein that allows a virus to replicate its genetic material. It is the only protein that all RNA viruses share, because it plays an essential role in how they propagate themselves. Each RNA virus, however, has small differences in the gene that codes for the protein that can help distinguish one type of virus from another.
So we screened a global database of RNA sequences from plankton collected during the four-year Tara Oceans expeditions global research project. Plankton are any aquatic organisms that are small to swim against the current. They’re a vital part of ocean food webs and are common hosts for RNA viruses. Our screening ultimately identified over 44,000 genes that code for the virus protein.
Our next challenge, then, was to determine the evolutionary connections between these genes. The more similar two genes were, the more likely viruses with those genes were closely related. Because these sequences had evolved so long ago (possibly predating the first cell), the genetic signposts indicating where new viruses may have split off from a common ancestor had been lost to time. A form of artificial intelligence called machine learning, however, allowed us to systematically organize these sequences and detect differences more objectively than if the task were done manually.